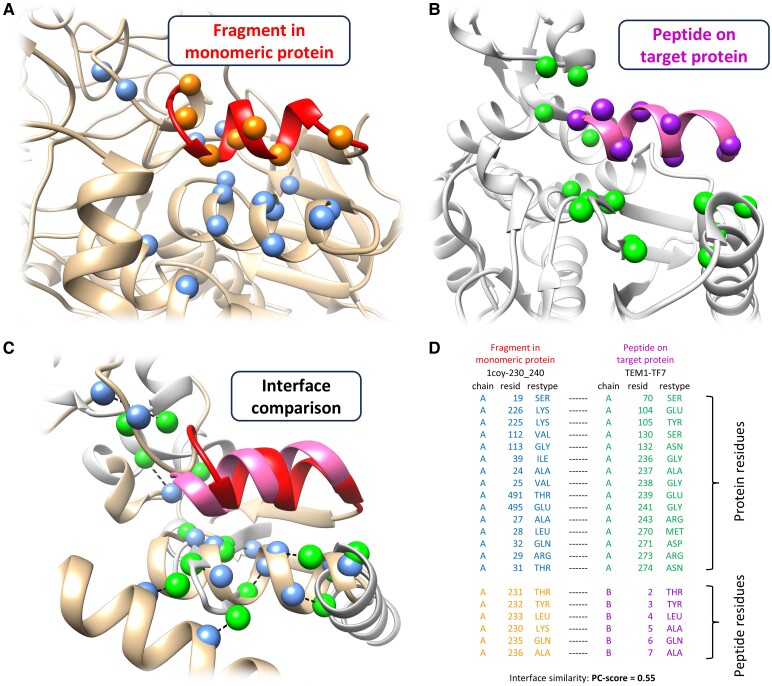
Fig. 1.
Illustration of the key idea of MDockPeP2_VS. A fragment derived from a monomeric protein (A) is highly likely to bind to a target protein (B) with a similar conformation when the interfacial residues are conserved (C and D). A) A fragment (residues 230–240, colored red) from a monomeric protein (PDB ID: 1coy). B) The binding mode of the peptide TF7 (highlighted in hot pink) on the target protein (TEM-1; PDB ID: 1s0w, chain A) predicted by the docking engine implemented in MDockPeP2_VS. TF7 shares the identical sequence with the red fragment displayed in (A). C and D) The two interfaces (A and B) are superimposed, and their physicochemical similarity is measured by the program PCalign (22). A PC score of 0 indicates no similarity, while a PC score of 1 reflects complete identity. The Cα atoms of conserved residues at the interface are shown as blue and green spheres in (A–C), respectively. Dashed lines in (C) indicate the correspondence between each conserved residue pair’s Cα atoms.