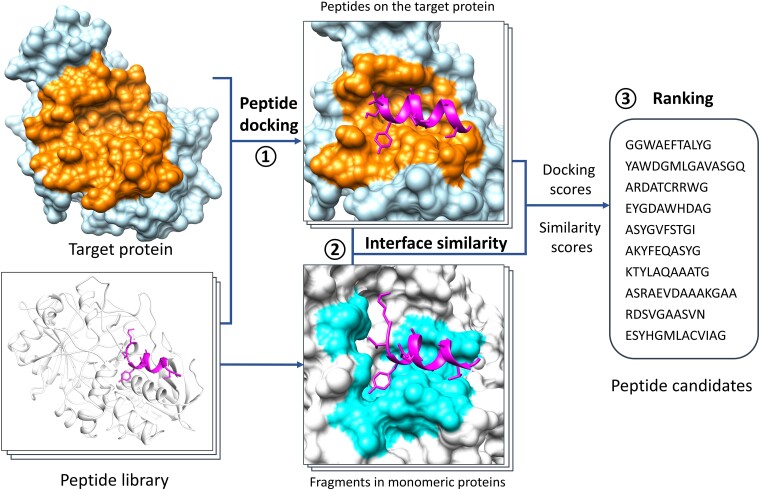
Fig. 2.
The flow chart of MDockPeP2_VS, which can be briefly divided into three steps. Step 1: protein–peptide docking. Peptides in a newly constructed peptide library are docked to a target protein using the program Vina_pep. The peptide library consists of fragments (10–15 amino acids) extracted from monomeric protein structures deposited in the PDB. Step 2: interface comparison. For each peptide, the predicted protein–peptide interacting interface is compared with the interface of the corresponding sequence fragment in the monomeric protein. Step 3: ranking. Peptides in the library are ranked by a hybrid scoring function, which combines protein–peptide docking scores and interface similarity scores. A few dozen peptide candidates are selected for synthesis and experimental validation. See the main text for details.