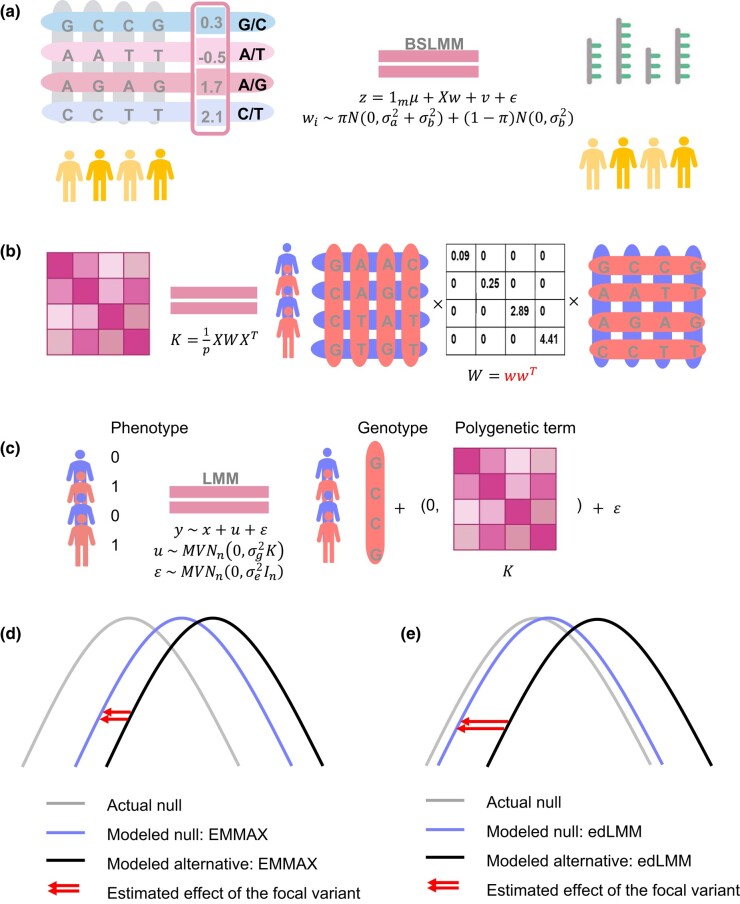
Fig. 1.
The edLMM protocol. a) The BSLMM tool (Bayesian sparse linear mixed model) is used to train the weights of each genetic variant, using genotype-expression data from a reference panel such as GTEx. b) The calculated weights are used to compute the weighted GRM in a GWAS dataset. c) The weighted GRM is used as the variance–covariance matrix of the multivariate normal distribution in an LMM for the GWAS. Panels d) and e) show the null and alternative distributions of genetic variants under EMMAX (the standard LMM) and edLMM protocol. The newly formed null distribution in panel e) is more accurate, signifying the statistical effect of the focal variant (red arrows).