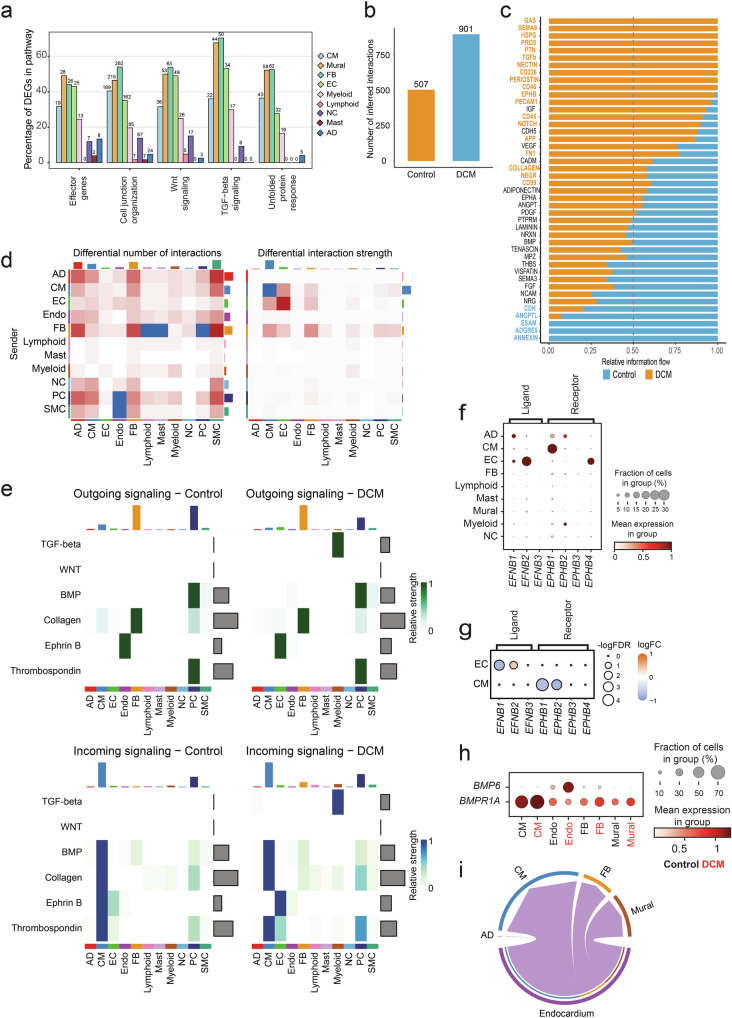
Extended Data Fig. 7. Intercellular interactions in DCM inferred from single nuclei transcriptomics.
a, Percentage of genes within candidate gene enriched pathways that are differentially expressed in DCM compared with controls, stratified by cell type. b, Total number of interactions between cell types in DCM (blue) and control (orange). c, Relative information flow of curated receptor-ligand intercellular, highlighting pathways that are significantly increased in DCM (orange) or control (blue). d, Heat map showing total overall differences in interaction number and strength between cell types (red – increased in DCM, blue – decreased). e, Heat map showing outgoing (green) and incoming (blue) signals for prioritized gene enriched pathways (TGF-beta and WNT pathways) and specific pathways of prioritised genes (BMP, Collagen, Ephrin B and thrombospondin). f, Expression levels of ephrin-B ligand and receptors across major cell types. Mean expression is scaled from minimum to maximum, and proportion of expressing nuclei within a cell type indicated by dot size. g, Increased expression of EFNB2 (ligand) in endothelial cells (EC) and decreased expression of EPHB1 (receptor) in cardiomyocytes (CM) in DCM. Dot colour represents change in expression compared with control, and dot size represents the FDR-adjusted P-value. h, Expression levels of BMP6 and BMPR1A in CM, endocardial, fibroblast (FB), and mural nuclei, stratified by HCM (red) and control (black) status. Mean expression is scaled from minimum to maximum, and proportion of expressing nuclei within a cell type indicated by dot size. i, Chord plot showing that majority of endocardial (purple) BMP6-BMPR1A signaling is to cardiomyocytes (blue), followed by mural (brown) and fibroblasts (orange). Dot colour reflects the communication probabilities and dot size represents P-values computed from one-sided permutation test. AD – adipocyte; CM – cardiomyocyte; EC – endothelial cell; Endo – endocardial cell; FB – fibroblast; NC – neuronal cell; PC – pericyte; and SMC – smooth muscle cell.