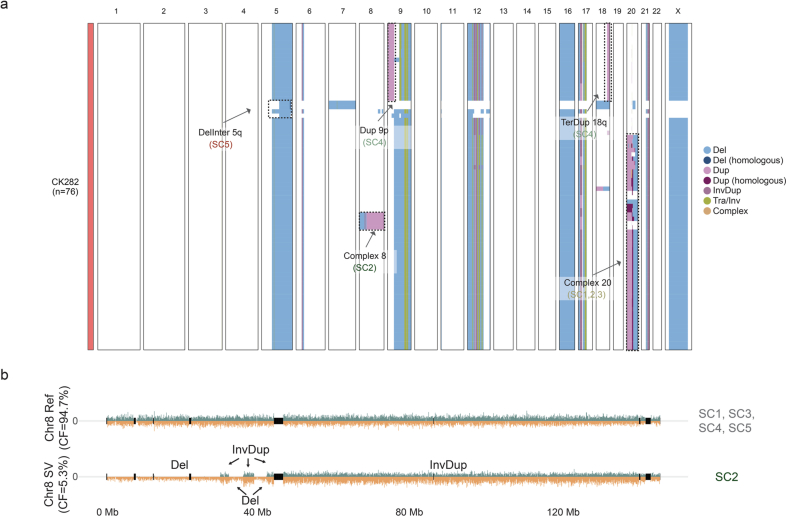
Extended Data Fig. 3. Subclonal heterogeneity in CK282.
a Karyotype heatmap of 76 single cells arranged using Ward’s method for hierarchical clustering of structural variant genotypes in CK282. Examples of subclone-specific structural variants are labelled in the heatmap. b Strand-specific read depth of two representative single cells from CK282 showing a normal chromosome 8 (reference, top) and a complex genetic rearrangement comprising of two inverted duplications (InvDups), three deletions (Dels) and one larger InvDup, spanning the whole chromosome 8 (bottom). Reads denoting somatic structural variants, discovered using scTRIP, mapped to the Watson (orange) or Crick (green) strand. Del: Deletion, Dup: Duplication, Inv: Inversion, Tra: Translocation, Inter: Interstitial, Ter: Terminal, Chr: Chromosome, CF: Cell fraction, SV: Structural variant.