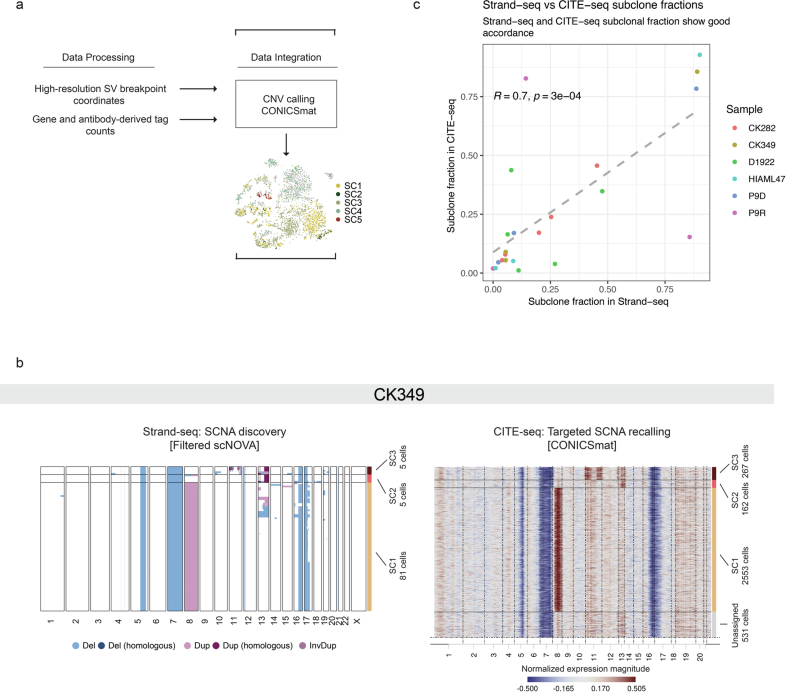
Extended Data Fig. 6. Integration of scNOVA with CITE-seq.
a Schematic of the data integration framework for scNOVA-CITE. Single-cell structural variant (SV) information from scTRIP and single-cell gene expression from CITE-seq was used as input for CONICSmat25, a computational tool for targeted somatic copy-number alteration (SCNA) recalling from scRNA-seq data. b SCNA discovery based on scNOVA from Strand-seq data (left) and targeted SCNA recalling based on CONICSmat from CITE-seq data (right) in patient CK349. Subclone assignments and corresponding cell numbers are shown on the right of each heatmap. c Subclone fraction in Strand-seq data vs. subclone fraction in CITE-seq data. Each dot represents a subclone and the dashed line shows the linear fit. Correlation was calculated using two-tailed Spearman correlation. CNV: Copy number variation, Del: Deletion, Hom: Homologous, Dup: Duplication, InvDup: Inverted Duplication.