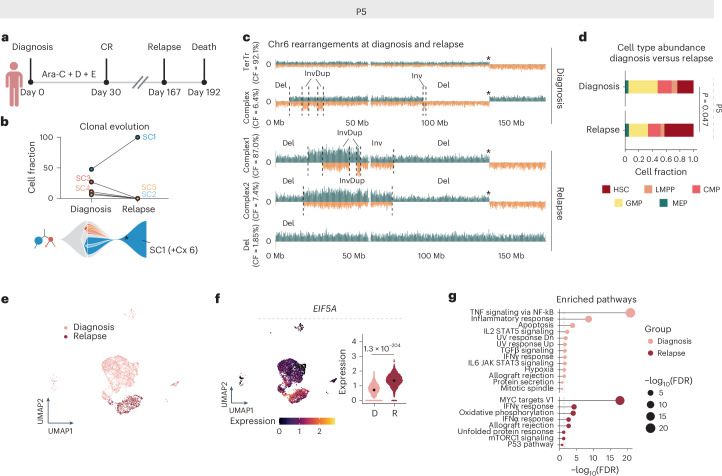
Fig. 6. Relapse is driven by a genetically evolving subclone in patient P5.
a, Disease timeline for patient P5. Panel a created with BioRender.com. b, Cell fraction of patient P5 subclones at diagnosis (D1922) and at relapse (R0836) based on the scTRIP data. The lines connect different time points (diagnosis versus relapse) of the same subclone (top). Fish plot (bottom) shows the inferred clonal evolution pattern and the subclonal tree the hierarchy of structural variant subclones at diagnosis, with the size of the circle relative to the clonal population. c, Depiction of example cells at diagnosis and relapse with differing rearrangements at chromosome 6. Asterisk denotes translocation breakpoint. d, Stacked bar plots showing the fraction of indicated HSPC-like states out of all cells at diagnosis and relapse. Cell types were annotated using a micrococcal nuclease (MNase)-seq reference dataset from index-sorted healthy CD34+ bone marrow cells and cell typing was pursued using scNOVA. The P value indicates the different abundance of HSC-like cells between the time points from two-sided Fisher’s exact test (nDiagnosis-HSC = 15 and nRelapse-HSC = 23, nDiagnosis-other = 48 and nRelapse-other = 31). e, Weighted nearest neighbor-based UMAP plots of diagnosis and relapse leukemic cells from patient P5 CITE-seq data. Cells are colored based on disease stage. f, Expression of EIF5A in single cells at diagnosis and relapse (nDiagnosis = 3,444 and nRelapse = 1,102). Beeswarm plots show the 95% CI for the mean and the gene expression comparison shows the Padj value from two-sided, pairwise Welch’s t-test. g, Enriched pathways at diagnosis and relapse. Genes with false discovery rate (FDR) < 0.05 and log(fold-change) > 0.25 were included in the analysis. CMP, common myeloid progenitor; CR, complete remission; Cx, complex; D, daunorubicin; E, etoposide; GMP, granulocyte–macrophage progenitor; TerTr, terminal translocation.