Fig. 1.
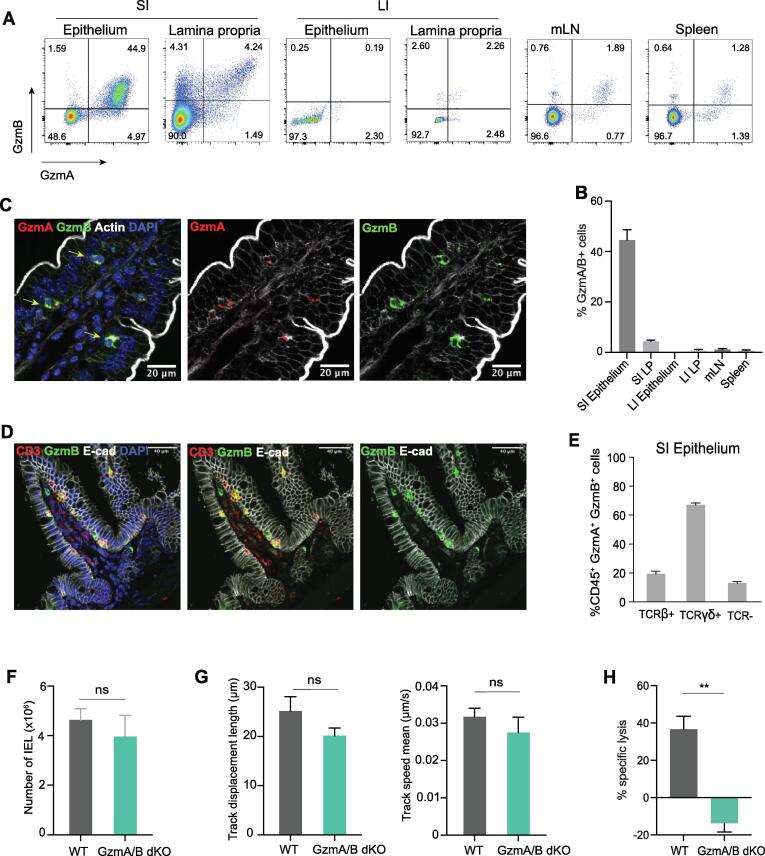
IEL are the main source of Gzms in the mouse at steady state. A. Representative flow cytometry dot plots of GzmA and GzmB expression in the small intestinal (SI) epithelium and lamina propria, large intestinal (LI) epithelium and lamina propria, mesenteric lymph nodes (mLN) and spleen of naïve WT mice (left). B. Bar graphs showing the quantification of GzmA+/GzmB+ cells in different organs shown in (A), n > 3 mice per organ. C. Immunofluorescent micrographs showing cells expressing GzmA (red) and GzmB (green) in the epithelial layer of a jejunal villus. Sections were counterstained with phalloidin to show actin (white) and DAPI (blue) to show nuclei. Scale bar = 20 µm. D. Immunofluorescent micrographs showing cells expressing CD3 (red), GzmB (green) and E-cad (white) in the jejunal villus. Scale bar = 40 µm. E. Bar graphs showing which CD45+ cell subsets express GzmA and GzmB in the epithelium (n = 5). F-H. Bar graphs showing F. CD45+ C103+ total IEL in co-housed WT and GzmA/B dKO (n = 5), G. Track displacement (left) and mean track speed (right) of WT and GzmA/B dKO IEL from mice which have been cohoused) in coculture with WT intestinal organoids (n = 3/group). H. Specific cell lysis of K562 cells by either WT or GzmA/B dKO IEL from mice that have been cohoused. IEL were cultured with K562 at a 40:1 ratio, in the presence of 20 ng/ml IL-15 and 1 μg/ml anti-CD3 (n = 3 per genotype). All data are presented as mean ± SEM. For (F-H), unpaired t-test was used to calculate the significance. ns: not significant, ** p < 0.01. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)