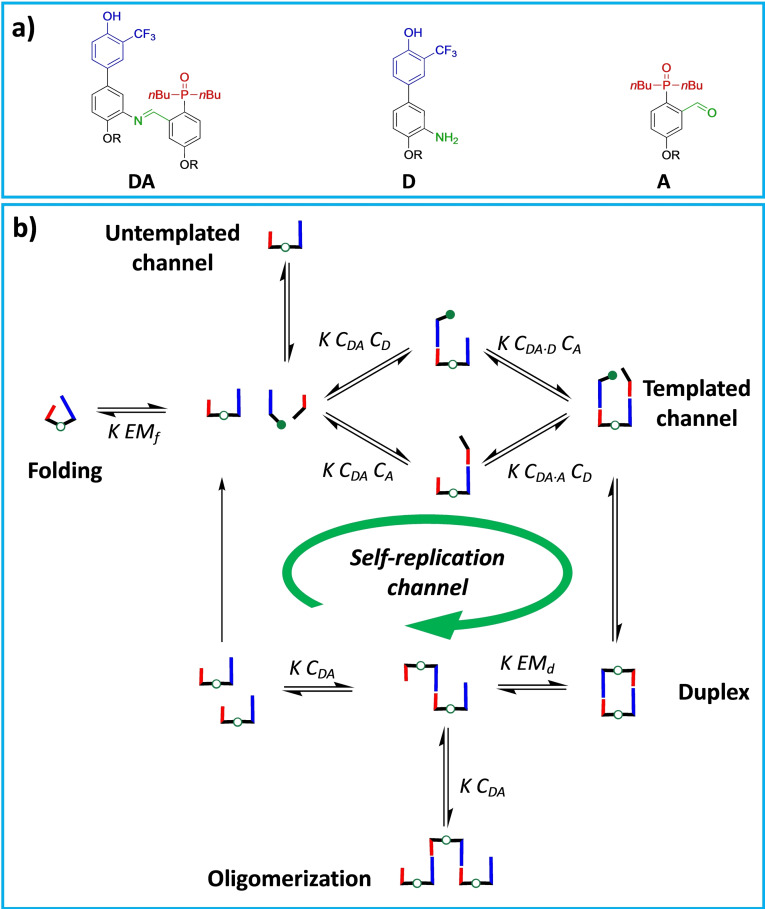
Figure 1.
a) Chemical structure of the recognition encoded molecules that compose the system: the monomers are equipped with hydrogen bond donor (trifluoromethylphenol, blue, for the aniline D) and acceptor (phosphine oxide, red, for the aldehyde A). Imine heterodimer DA is shown, and the linker chemistry is highlighted in green. R = 2‐ethylhexyl. b) Cartoon showing the pathways involved in this minimal replicating system: the imine template can be formed through a non‐templated channel and via self‐replication. The self‐assembly channels involving the imine dimer DA are highlighted: the first base‐pairing interaction can take place in an intramolecular fashion, leading to the 1,2‐folding path, or intermolecularly with a DA dimer or D/A monomers, leading respectively to the duplex channel or supramolecular oligomerization (template inhibition), or to the trimeric complex (templated channel). The outcome depends on the concentrations of the components C, the association constant for the intermolecular base‐pairing interaction K, and the effective molarities for folding (EMf ) and duplex formation (EMd ).