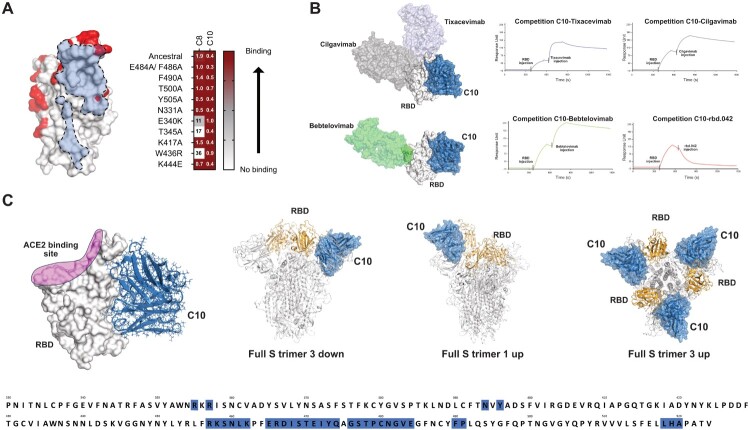
Figure 3.
Docking model of C10 mAb binding to Spike RBD. (A) Binding capacity of C8 and C10 mAb to SARS-CoV-2 mutated RBDs shown as heatmap (right; EC50 values are reported in nanomolar). C10 fingerprint on RBD with mutated residues tested in binding assays (left). Residues mutated in the RBDs tested are shown in red. Light blue is the C10 fingerprint (6Å). (B) C10 mAb competition with clinically previously used mAbs and the neutralizing mAb rbd.042 for RBD binding (right). C10 antibody was immobilized on chip; RBD was then flowed to form the RBD/antibody complex, and shortly thereafter, Tixacevimab, Cilgavimab, Bebtelovimab or rbd.042 mAbs were injected. The binding event detected at the final step for Tixacevimab, Cilgavimab and Bebtelovimab, indicates that these second antibodies injected have a different epitope compared to the first (immobilized) antibody; instead, no binding, meaning competition, is detected for rbd.042. Structural representation of RBD with bound C10 and different Abs used for cross-competition experiments (left). (C) RBD surface representation with bound C10 based on the obtained with the docking model (left). C10 is shown in blue and ACE2 binding site is shown on RBD in pink. The putative epitope recognized by C10 (blue) encompasses amino acid residues dispersed across various regions within the RBD, predominantly situated in a region near the Subdomain 1 (SD1). C10 epitope accessibility on Spike trimer based on the docking model (right).