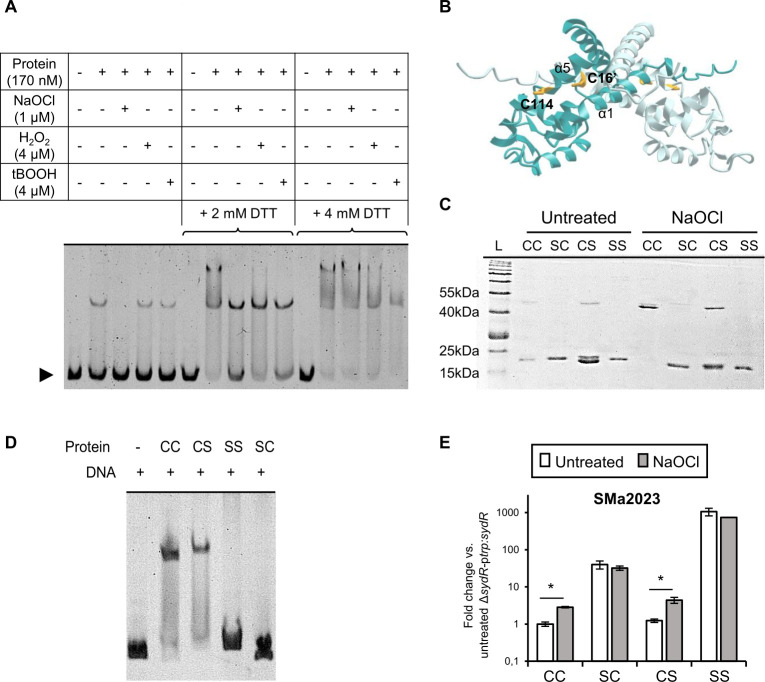
Fig 3.
SydR-binding activity is inhibited by oxidation and depends on the redox-active cysteine C16. (A) EMSA with DNA (sydR-SMa2023 intergenic region) SydR′ in 0.5 mM DTT buffer treated or not with various oxidants was performed at 25°C for 25 min, followed by an additional 25 min incubation with 2 or 4 mM DTT as indicated. (B) Three-dimensional structural model of SydR in homodimeric configuration, generated using AlphaFold Protein Structure Database (https://colab.research.google.com/github/sokrypton/ColabFold/blob/main/AlphaFold2.ipynb). One subunit is colored turquoise-blue, and the other subunit is colored pale blue. Cysteines are shown as yellow sticks. C16 is predicted to be located at the dimer interface, and C114 to be exposed at the surface, like C22 and C127 in Xanthomonas campestris OhrR (25). Non-reducing SDS-page (C) and EMSA (D) using SydR′ WT protein (CC) and mutant variants harboring the C16S (SC), C114S (CS), and C16S&C114S (SS) mutations, treated with DTT then, when indicated, with NaOCl. (E) RT-qPCR analysis of SMa2023 expression in ΔsydR-ptrp:sydR strain and ΔsydR derivatives encoding SydR mutant variants (SC, CS, or SS). The values shown are the means ± SEM of three independent experiments. Student’s t test was used to assess the statistical significance (*P < 0.05).