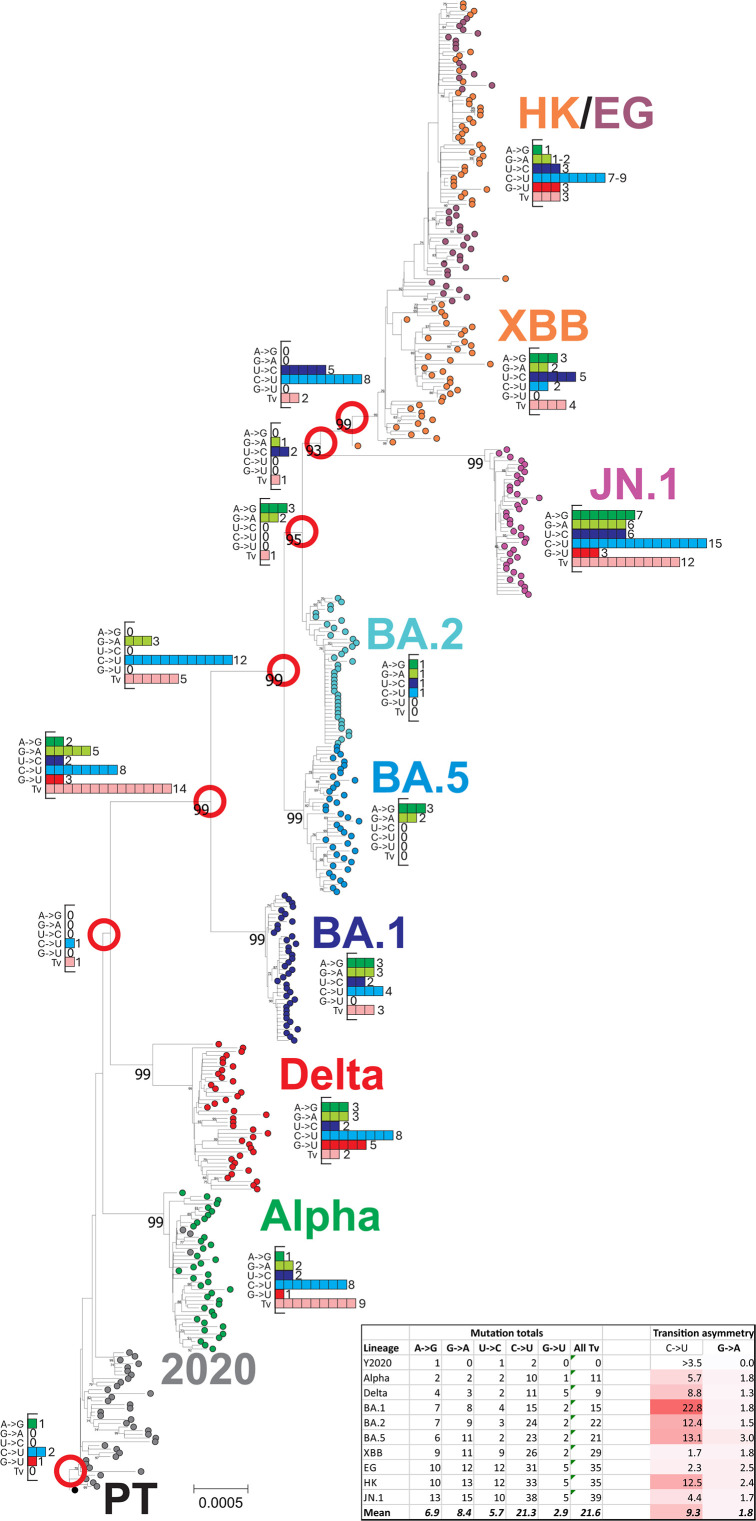
Fig 1.
Sequence changes in the emergence of SARS-CoV-2 lineages. Phylogenetic tree of coding region sequences of lineages of SARS-CoV-2 sequentially emerging during the course of the COVID-19 pandemic during 2020–2024. Inset histograms show the numbers of each transition, G→U, and total transversions that occurred between nodes (red circles) and descendant nodes or consensus lineage sequence. Substitution totals for each lineage are provided in the inset table, along with estimated transition asymmetries that account for base composition. The tree was constructed by neighbor joining of Jukes–Cantor corrected distances; robustness of grouping was determined by bootstrap re-sampling of 100 pseudo-replicas; support values of >70% are shown on the branches.