FIGURE 1.
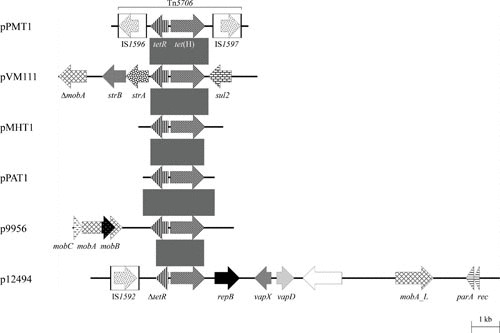
Schematic representation of the structure and organization of genes found in tet(H)-carrying plasmids from P. multocida, M. haemolytica, [P.] aerogenes, and A. pleuropneumoniae. Comparison of the maps of the partially sequenced plasmids pPMT1 (accession no. Y15510) and pVM111 (accession nos. AJ514834 and U00792), both from P. multocida, pMHT1 (accession no. Y16103) from M. haemolytica, and pPAT1 (accession no. AJ245947) from [P.] aerogenes (accession no. Z21724) and the completely sequenced plasmids p9956 (accession no. AY362554; 5,674 bp) and p12494 (accession no. DQ517426; 14,393 bp), both from A. pleuropneumoniae. Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: tetR-tet(H) (tetracycline resistance), sul2 (sulfonamide resistance), and strA and strB (streptomycin resistance); plasmid replication: repB; mobilization functions: mobA, mobB, mobC, and mobA_L; recombination functions: rec; DNA partition: par; virulence: vapD and vapX; unknown function: the open reading frame indicated by the white arrow. The Δ symbol indicates a truncated functionally inactive gene. The white boxes in the maps of pPMT1 and p12494 indicate the limits of the insertion sequences IS1592, IS1596, and IS1597; the arrows within these boxes indicate the reading frames of the corresponding transposase genes. Gray shaded areas indicate the tetR-tet(H) gene region common to all these plasmids with ≥95% nucleotide sequence identity. A distance scale in kilobases is shown at the bottom of the figure.
