FIGURE 3.
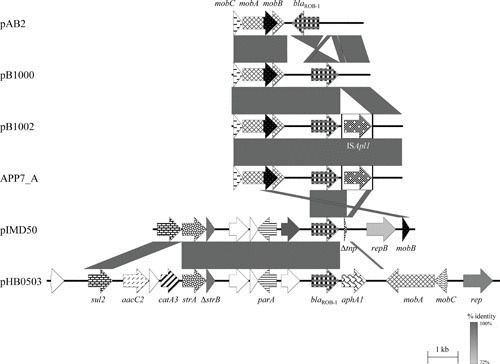
Schematic representation of the structure and organization of the blaROB-1-carrying resistance plasmids from M. haemolytica, [H.] parasuis, P. multocida, A. pleuropneumoniae, and “A. porcitonsillarum.” Comparison of the maps of blaROB-1-carrying resistance plasmids pAB2 (accession no. Z21724; 4,316 bp) from M. haemolytica, pB1000 (accession no. DQ840517; 4,613 bp) from [H.] parasuis, pB1002 (accession no. EU283341; 5,685 bp) from P. multocida, APP7_A (accession no. CP001094; 5,685 bp) from A. pleuropneumoniae, pIMD50 (accession no. AJ830711; 8,751 bp) from “A. porcitonsillarum,” and pHB0503 (accession no. EU715370; 15,079 bp) from A. pleuropneumoniae. It should be noted that another three pIMD50-related blaROB-1-carrying resistance plasmids from “A. porcitonsillarum” have been sequenced completely: pKMA5 (accession no. AM748705), pKMA202 (accession no. AM748706), and pKMA1467 (accession no. AJ830712). Genes are shown as arrows, with the arrowhead indicating the direction of transcription. The following genes are involved in antimicrobial resistance: sul2 (sulfonamide resistance), strA and strB (streptomycin resistance), blaROB-1 (β-lactam resistance), aacC2 (gentamicin resistance), catA3 (chloramphenicol resistance), and aphA1 (kanamycin/neomycin resistance); plasmid replication: rep; mobilization functions: mobA, mobB, and mobC; resolvase function: res; DNA partition: parA; unknown function: open reading frames indicated by white arrows. The prefix Δ indicates a truncated functionally inactive gene. Gray-shaded areas indicate the regions common to plasmids, and the different shades of gray illustrate the percentages of nucleotide sequence identity between the plasmids, as indicated by the scale at the bottom of the figure. A distance scale in kilobases is shown.
