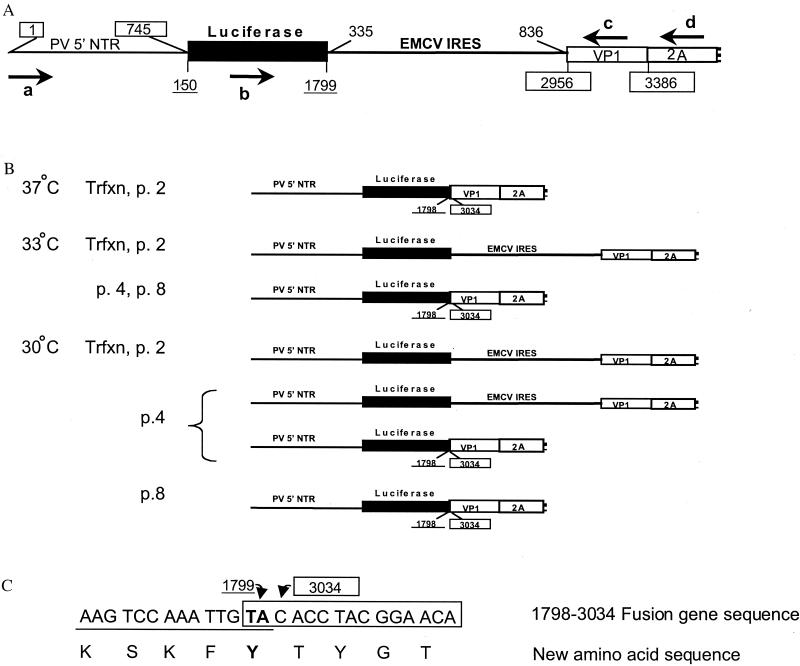
FIG. 3.
Analysis of PV-Luc-EMCV genomes following transfection and serial passage. (A) Schematic of the starting dicistronic genome. The boxed numbers refer to nucleotides in the PV genome. The underlined numbers refer to positions in the gene encoding luciferase. Nucleotides 335 to 836 of the EMCV genome correspond to the IRES [without poly(C) tract]. Primers used for RT-PCR (a to d) are marked by arrows and described in Materials and Methods. (B) Schematic of the major genomes recovered by RT-PCR from transfections and serial passages. The RT-PCR products recovered from transfections (Trfxn) or serial passages in the presence of VV-P1 (p2, p4, and p8) were cloned and sequenced. The major deleted genomes recovered at all temperatures consisted of a fusion between the luciferase (nucleotide 1798) and VP1 (nucleotide 3034) genes to maintain the translational reading frame. The original dicistronic genome was also recovered from early passages at lower temperatures. (C) Nucleotide and predicted amino acid sequences of the luciferase-VP1 gene fusion. The entire EMCV IRES was deleted from the original dicistronic construct, along with 78 nucleotides of the PV VP1 coding region (from nucleotides 2956 to 3033). The PV nucleotides are boxed, and the luciferase nucleotides are underlined. The first two nucleotides of the luciferase stop codon are in bold; the last two luciferase nucleotides and the first two PV nucleotides are identical. Note that although a Y-G amino acid dipeptide is present a few amino acids from the 3′ end of the luciferase gene, the P4 amino acid is F rather than the L required for optimal 2Apro cleavage (9).