Fig. 1 |. Schematic of non-B DNA structures.
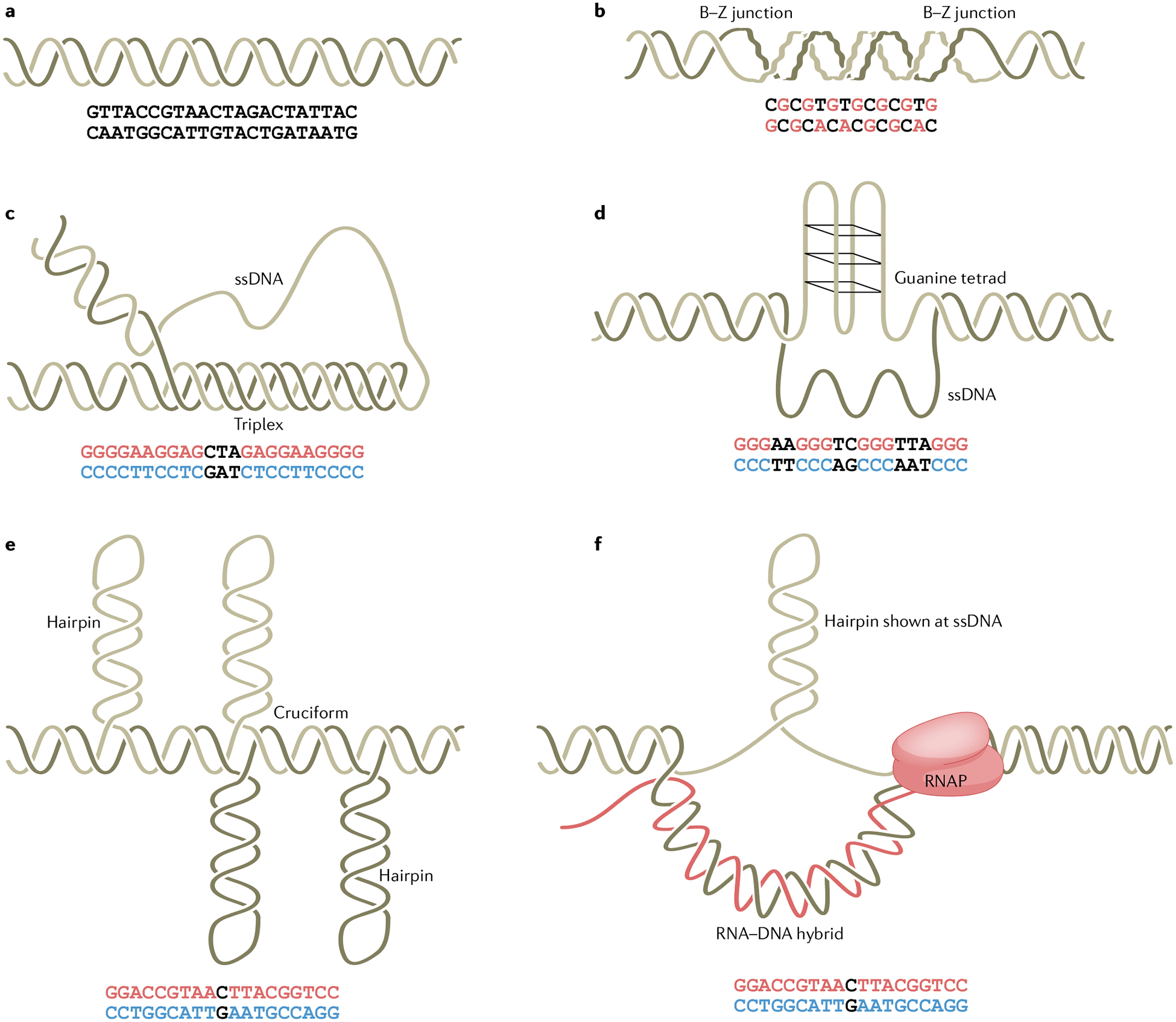
a, Canonical B-form DNA. b, Z-DNA forms at alternating purine–pyrimidine sequences, where the syn-formation purines and anti-conformation pyrimidines twist the backbone into a zigzag shape310. c, H-DNA forms at polypurine or polypyrimidine sequences that contain a mirror repeat, where half of the repeat in single-stranded form folds back into the major groove of the DNA duplex to form a triplex structure via Hoogsteen hydrogen bonding311,312. H-DNA can exist in various isomers depending on strand orientation and whether the purine-rich or pyrimidine-rich strand is used as the third strand. d, G quadruplexes form at sequences containing four runs of three or more guanines. Four guanine bases associate through Hoogsteen hydrogen bonding (guanine tetrad), and three continuous guanine tetrads stack to form a G quadruplex (G4 DNA)88,313. e, Cruciform or hairpin structures form at inverted-repeat sequences167,314,315, whereby two symmetrical arms self-anneal to form a duplex stem. f, R-loops contain a nascent RNA strand annealed to the DNA template strand316, leaving the non-template strand unpaired, which can adopt a stable structure, such as a hairpin or G4 DNA. The red/blue letters in the sequences represent the bases involved in the non-B conformation. RNAP, RNA polymerase; ssDNA, single-stranded DNA.
