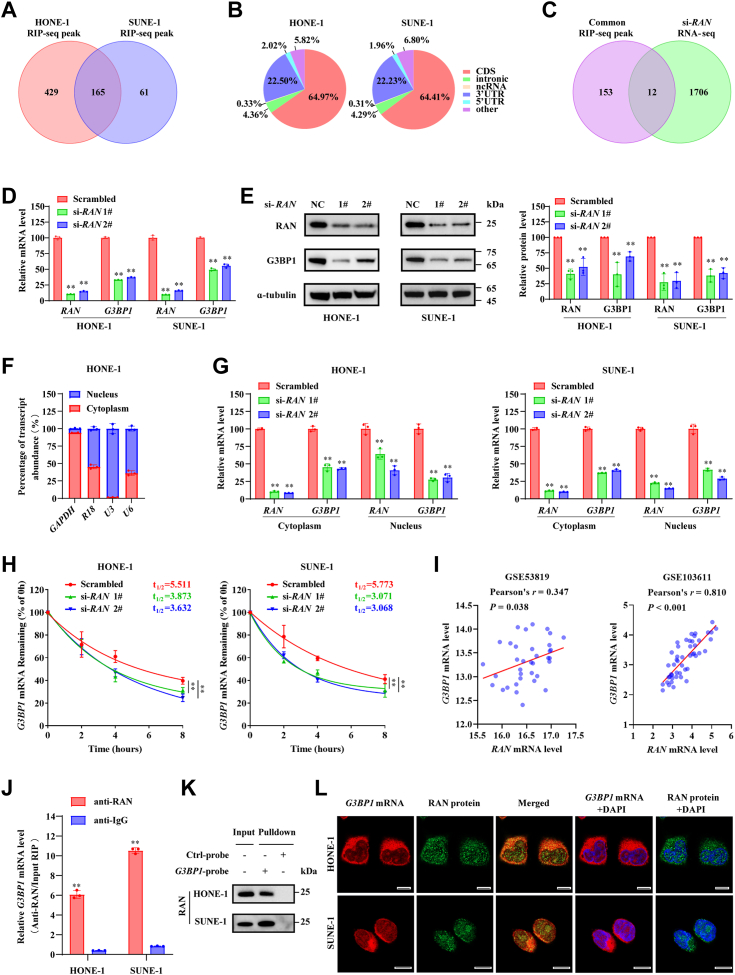
Figure 4.
RAN directly binds and stabilizes G3BP1 mRNA.A, Venn diagram of genes identified by RIP-seq in HONE-1 or SUNE-1 NPC cell lines (logOddScore >1). B, pie charts showing the distribution of reads recognized by RIP-seq on gene functional elements. C, venn diagram of differentially expressed genes identified by RNA-seq after RAN silencing and overlapping genes identified by RIP-seq. D, relative levels of G3BP1 mRNA level with or without RAN silencing were detected by RT-qPCR. Data are presented as the mean ± SD (n = 3). E, G3BP1 protein level with or without RAN silencing was detected by Western blotting. The blots are the representation of three independent experiments. Data are presented as the mean ± SD (n = 3). F, nuclear/cytosol RNA fractionation assays were used to identify the suitable internal control. Data are presented as the mean ± SD (n = 3). G, relative levels of G3BP1 mRNA level in cytoplasm or nucleus, which were normalized to GAPDH or U3, was indicated by RT-qPCR upon knockdown of RAN in HONE-1 and SUNE-1 cells. Data are presented as the mean ± SD (n = 3). H, after treatment with actinomycin D (10 μg/ml), G3BP1 mRNA level was quantified at indicated times in control and RAN-silenced cells. The half-life of G3BP1 mRNA was analyzed by plotting degradation curves. Data are presented as the mean ± 95%CI (n = 3). I, Pearson correlation analysis of RAN and G3BP1 levels in different GEO databases (GSE53819 and GSE103611). J, relative enrichment of G3BP1 mRNA immunoprecipitated by anti-RAN or anti-IgG antibody was indicated by RT-qPCR. Data are presented as the mean ± SD (n = 3). K, enrichment of RAN proteins pulled down by biotin-labeled G3BP1 probes from in vitro transcription or control antisense probes was detected by Western blotting. L, RAN protein distribution in cells was recognized by IF (green), G3BP1 mRNA distribution in cells was recognized by FISH (red), and cell nuclei were stained with DAPI (blue). The scale bar represents 10 μm. ∗p < 0.05 and ∗∗p < 0.01. The significant differences were assessed using one-way ANOVA (D, E, and G), two-way ANOVA (H), and t test (J). The significant differences in correlations were assessed using the Pearson correlation analysis (I). GEO, Gene Expression Omnibus; RT-qPCR, reverse transcription quantitative real-time PCR; NPC, nasopharyngeal carcinoma; DAPI, 4′, 6-diamino-2-phenylindole; IF, immunofluorescence; CI, confidence interval.