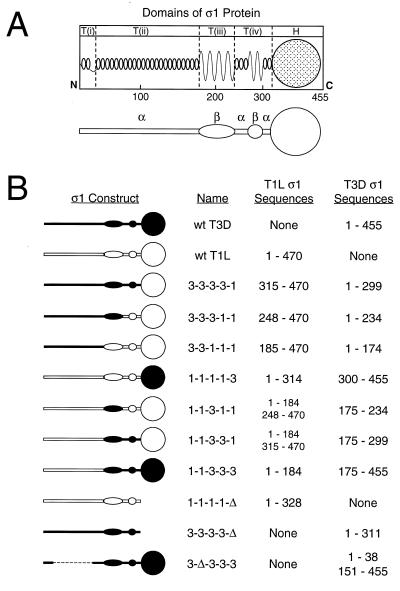
FIG. 1.
Chimeric and truncated ς1 proteins used for studies of carbohydrate binding. (A) Model of ς1 structure depicting predicted secondary structures and correlating primary amino acid sequence with morphologic regions of ς1 (T(i), T(ii), T(iii), T(iv), and H) seen in computer-processed electron micrographic images of ς1 protein isolated from virions (16, 31). In the simplified version of this model shown below, α-helical regions of the tail domain are indicated by horizontal bars and regions of β-strand/β-turn are symbolized by ovoid shapes. The globular head domain (H) is depicted as a circle. (B) Sequence features of chimeric and truncated ς1 constructs. White symbols represent sequences derived from T1L ς1, and black symbols represent sequences derived from T3D ς1. Constructs are named according to the parental origin of ς1 morphologic regions as previously described (16, 31): 1, sequences derived from T1L ς1; and 3, sequences derived from T3D ς1. Sequences corresponding to morphologic regions T(i), T(ii), T(iii), T(iv), and H are represented by the first, second, third, fourth, and fifth characters, respectively, of construct names. Δ, deleted sequences. The sequences of T1L and T3D ς1 comprising each construct are denoted by numbers corresponding to ς1 amino acid residues reported by Nibert et al. (31) (T1L) and Bassel-Duby et al. (2) (T3D).