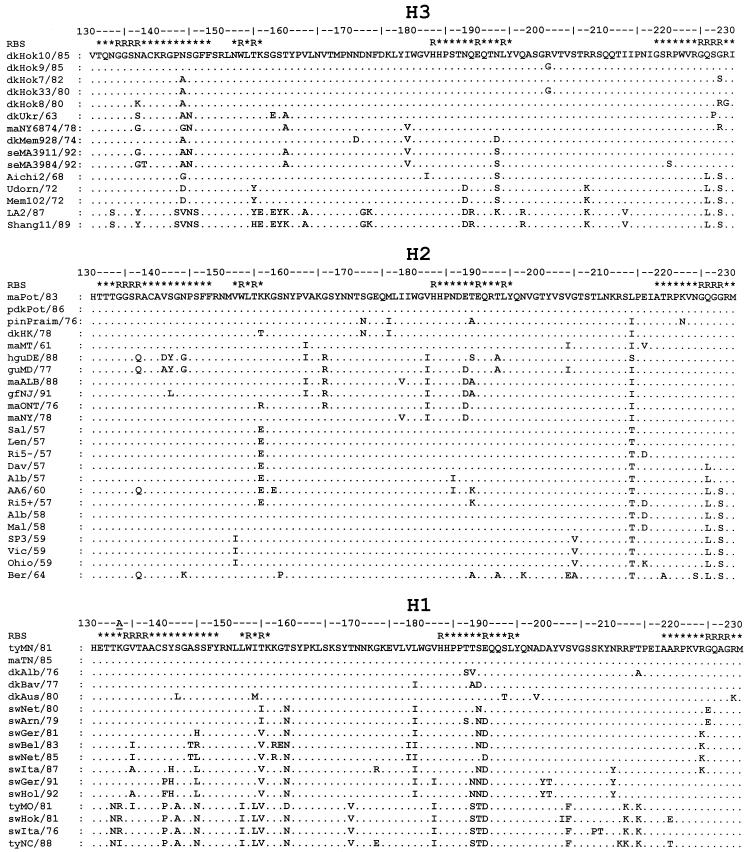
FIG. 2.
Partial HA amino acid sequences (HA1 positions 130 to 230) of influenza viruses that were tested for their binding to sialylglycopolymers. Full names of the virus strains are listed in Tables 1 to 3. Differences with respect to the top sequence are shown. The H3 numbering system is used; the position in the H1 HA that is absent from the H3 HA is indicated by an underscore. The RBS line shows the position of the amino acid with respect to the HA receptor-binding site. The “R” designates that the amino acid residue contacts either sialic acid or penultimate galactose in the X31 virus HA complex with 3′-sialyllactose (1HGG structure; Brookhaven Protein Database). The star indicates that an amino acid is within 15 Å of the C2 carbon atom of the sialic acid in the 1HGG structure. The figure was generated with GeneDoc 2.3 software (25).