Abstract
Identifying residues critical to protein–protein binding and efficient design of stable and specific protein binders are challenging tasks. Extending beyond the direct contacts in a protein–protein binding interface, our study employs computational modeling to reveal the essential network of residue interactions and dihedral angle correlations critical in protein–protein recognition. We hypothesized that mutating residues exhibiting highly correlated dynamic motion within the interaction network could efficiently optimize protein–protein interactions to create tight and selective protein binders. We tested this hypothesis using the ubiquitin (Ub) and MERS coronaviral papain-like protease (PLpro) complex, since Ub is a central player in multiple cellular functions and PLpro is an antiviral drug target. Our designed ubiquitin variant (UbV) hosting three mutated residues displayed a ~3,500-fold increase in functional inhibition relative to wild-type Ub. Further optimization of two C-terminal residues within the Ub network resulted in a KD of 1.5 nM and IC50 of 9.7 nM for the five-point Ub mutant, eliciting 27,500-fold and 5,500-fold enhancements in affinity and potency, respectively, as well as improved selectivity, without destabilizing the UbV structure. Our study highlights residue correlation and interaction networks in protein–protein interactions, and introduces an effective approach to design high-affinity protein binders for cell biology research and future therapeutics.
Keywords: dihedral angle correlation network, protein-based inhibitors, ubiquitin variant, papain-like protease, nanomolar binder
Introduction
Understanding what drives protein–protein binding and selecting appropriate protein residues for modification to strengthen protein–protein interactions (PPIs) are crucial to designing a protein binder that targets its binding partner.1,2 Strategies that can efficiently and accurately identify residues to enhance PPIs have broad applications in therapeutics and studies of cell biology. Knowledge-based, physics-based, and data-driven methods have all been developed previously to explore PPIs and to select mutations that enhance them.3–9 Computational and combinatorial libraries or in vitro evolutionary approaches also represent popular protein engineering strategies to design stable and specific protein binders.10–12 Importantly, minimizing the number of residues mutated to significantly enhance PPIs lowers the possibility of engineering unstable proteins. Nevertheless, protein engineering remains challenging, as highly integrated molecular modeling and experimental techniques are needed to understand PPIs in order to re-engineer a protein to increase its binding affinity.
PPI networks are extremely complex, so selecting an appropriate target system for experimental modification requires specialized expertise. Here, we chose Ubiquitin (Ub) as our target system, as it plays critical roles in numerous biological functions.13 Ub is a small 76-residue protein associated with post-translational modifications. This regulatory protein canonically binds to its cascade E1-E2-E3 enzymes to drive ubiquitination and Ub chain formation, thereby modifying nearly half of the human proteome.14,15 Conversely, deubiquitinases (DUBs)16 cleave the covalent isopeptide bonds from Ub chains or substrates to release Ub and its substrates. The interactome of Ub and cellular proteins have been assessed15 revealing that precise Ub network regulation governs cellular fates. Misregulation of the responsible enzymes significantly impacts cellular functions, leading to diseases such as cancers.17 Moreover, viral DUBs have been found to interfere with host antiviral defenses. For example, the Papain-like protease (PLpro) of coronaviruses (CoVs) is classified as a viral DUB specific to Ub and Ub-like ISG15.18,19 Previous studies have shown that PLpro alters host innate immune responses, which contributes to the rapid spread of CoVs (such as MERS, SARS-CoV, SARS-CoV-2),20–24 thus causing pandemics, mortality, and perturbing the global economy.25
The PLpro proteins of MERS and SARS-CoV-2 are crucial for viral replication through their role in proteolytic cleavage of viral nonstructural proteins (NSPs). The PLpro domain resides in NSP3, which drives viral genome replication and subgenomic RNA synthesis.26,27 PLpro recognizes and cleaves the NSP1–2, NSP2–3 and, NSP3–4 junctions after the amino acid sequence LXGG to yield functional viral proteins, as well as to perform deubiquitination and deISGylation.21,23,24 Deubiquitination and deISGylation alter host signaling pathways critical to induction of cellular antiviral and pro-inflammatory innate immune responses, ultimately suppressing the host antiviral response.21,28 Therefore, inhibiting PLpro simultaneously disrupts viral replication and prevents PLpro from impairing the innate immune response. Given both these properties, PLpro represents an ideal antiviral drug target.
Importantly, wild-type Ub (wtUb) exhibits high thermostability , so it is an ideal template for protein design. Re-engineered Ub also has potential advantages, such as enhanced binding specificity to PLpro and easier synthesis compared to chemical compounds. Screening of phage-displayed Ub variants (UbVs) against cognate enzymes, including MERS PLpro, has previously demonstrated the feasibility of regulating the activities of E3 ligases and DUBs.29–31 The phage-display screening technique focused on three surface patches in Ub to iteratively mutate and select variants that displayed tight binding. The resulting DUB UbVs proved to be strong inhibitors, exhibiting IC50 values in the range of 1–30 nM.30–32 As an alternative approach, computational data were used to rationally design a screening library for the identification of tightly binding regulatory UbVs for USP733 and USP21.34 A combined computational and phage-display screening of UbVs targeting USP7 resulted in an equilibrium constant (KD) for the U7Ub25.2540 variant of 56 nM, whereas for wtUb-USP7 it was >200 μM. A pool of 6,000 designed UbVs for USP21 revealed that ~10% of the variants tightly bound USP21 consistently between experimental and computational screenings. However, in silico screenings of such large UbV-USP21 pairings require intensive computational resources. Such expensive and time-consuming empirical screenings impede rational design of protein-based inhibitors.
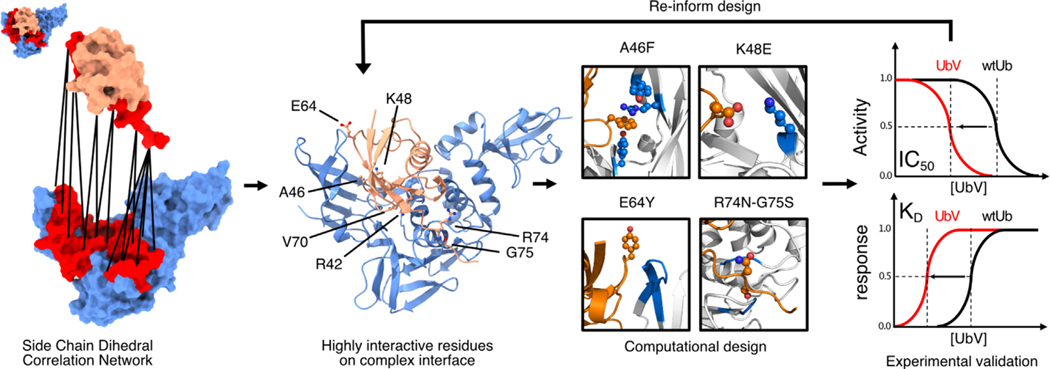
Here, we present an integrated computational and experimental approach to identify critical regions for protein–protein binding that display highly correlated dynamic motion. Specifically, we focus on side-chain dihedral angle correlations at the protein–protein contact interface where mutation of highly correlated residues resulted in both local and distal conformational changes. We demonstrate that mutating residues in these regions can efficiently optimize PPIs to create tight and selective protein binders. We show that our designed UbVs hosting two or three mutated residues achieved 3,500-fold inhibitory efficiency and binding affinity relative to wtUb for MERS PLpro (Table 1). MERS PLpro cleaves both K48- and K63-linked Ub chains,18,23 and it exhibits distinct inhibitor recognition specificity to that of the PLpro of SARS-CoV and SARS-CoV-2.35 We used non-covalent amino acid interaction and side-chain dihedral angle networks of the Ub and MERS PLpro (Ub-PLpro) complex to guide our design of UbVs that enhance UbV–PLpro binding affinity, thereby inhibiting PLpro activity. Initially, we designed two-point mutations for cost efficiency and to retain intact the overall complex structure. Integrating experimental data and computational analyses informed our experimental design to yield more UbVs (Fig. 1). Binding affinity KD and IC50 measurements of our designed UbVs support that more extensively mutated UbV3, UbV4, and UbV5 represent strong inhibitors.
Table 1.
Computational and experimental evaluation of the binding affinity between MERS PLpro and UbVs.
| Ubiquitin and variants | Binding energy (kcal/mol) | Dissociation time (ns) | IC50 | KD | PLpro selectivity* |
|
|---|---|---|---|---|---|---|
| MERS | SARS-CoV-2 | |||||
|
| ||||||
| wtUb | −41.95 ± 2.56 | 32 | 52.91 ± 6.98 μM | 40.75 ± 3.82 μM | − | − |
| A46F | −48.25 ± 1.09 | 67.5 | 1.64 ± 0.03 μM | 2.74 ± 0.29 μM | +++ | − |
| K48E | −49.52 ± 2.56 | 75 | 3.94 ± 0.42 μM | 5.15 ± 0.71 μM | ++ | − |
| E64Y | −42.00 ± 1.49 | 68 | 0.29 ± 0.03 μM | 0.46 ± 0.04 μM | ++++ | − |
| A46F-K48E (UbV2) | −50.54 ± 2.35 | 207 | 0.20 ± 0.00 μM | 0.22 ± 0.03 μM | +++++ | − |
| A46F-K48L | −49.03 ± 2.33 | 200 | 0.23 ± 0.01 μM | N/A | N/A | N/A |
| A46F-K48S | −47.13 ± 2.84 | N/A | 0.18 ± 0.01 μM | N/A | N/A | N/A |
| A46F-K48E-G75R | −45.24 ± 1.48 | 40 | 9.74 ± 0.15 μM | 11.41 ± 1.26 μM | N/A | N/A |
| A46F-K48I | N/A | N/A | 0.49 ± 0.04 μM | N/A | N/A | N/A |
| A46F-K48E-E64Y (UbV3) | −53.77 ± 1.37 | >250 | 14.84 ± 1.44 nM | 2.77 nM | ++++++ | − |
| A46F-K48E-R74N-G75S (UbV4) | −50.18 ± 0.88 | 65 | 0.11 ± 0.01 μM | 0.15 ± 0.02 μM | +++++ | − |
| A46F-K48L-R74N-G75S | −46.49 ± 0.98 | >250 | 0.13 ± 0.01 μM | 0.14 ± 0.02 μM | N/A | N/A |
| A46F-K48E-E64Y-R74N-G75S (UbV5) | −50.81 ± 3.25 | 88 | 9.71 ± 0.74 nM | 1.48 nM | +++++ | − |
| ME.2** | −47.98 ± 1.72 | >250 | 15.62 ± 2.54 nM | 53.2 ± 2.2 nM | +++++ | − |
| ME.4** | N/A | N/A | 28.57 ± 1.94 nM | 35.9 ± 1.6 nM | +++++ | − |
Figure 1.
Workflow for the rational design of ubiquitin (Ub) variants. Molecular modeling by means of molecular dynamics (MD) simulations and post-hoc analysis reveals highly correlated regions in the interface (red) between PLpro (blue) and Ub (orange). The workflow shows how we integrated computational and experimental design, validation, and interpretation.
Results and Discussion
The process of optimizing PPIs involves identifying residues suitable for mutation, and then determining substitutions aimed at generating novel protein variants. We used Molecular Dynamics (MD) simulation to reveal residues in protein interfaces displaying strongly correlated dynamic motions. The central hypothesis underlying our rationale is that residues exhibiting a high degree of correlation play a pivotal role within the protein–protein interface. Moving just one protein side chain can influence the motion of other side chains. Thus, mutating highly correlated residues not only can strengthen or weaken local interactions, but can also maximize the impact of changes across all correlated residues in the interface. Mutating these dynamically correlated residues can enhance intermolecular attractions and thereby significantly improve PPIs. We employed a dihedral angle correlation network to identify important protein residues in the PPI interface of Ub and MERS PLpro. This network allowed us to understand how different parts of the proteins move in relation to one another. Then, we selected candidate residues for mutation by means of a pairwise force distribution analysis. For each of our designed UbVs, we computed their binding energy data using molecular mechanics Poisson-Boltzmann surface area (MM/PBSA) and the dissociation time by means of PPI Gaussian-accelerated MD (PPI-GaMD). To further validate our results, we employed a fluorescence polarization (FP)-based inhibition assay to quantify their binding affinities and selectivities, and adopted circular dichroism (CD) spectroscopy to assess protein thermostability.
Identifying key residues in the PPI interface
Mutating highly correlated residues to increase overall binding affinity necessitates investigating localized attraction, structural dynamics, and the biological effects of protein residues. The process of mutant design involved initial adjustments of Ub residues within stable Ub-PLpro complex domains, such as the hydrophobic core for the S1 Ub (HCS1) and alpha helix in the canyon (AHC), before extending modifications to more flexible domains such as the finger-Ub interaction (FBI) and Ub chain cleavage (UCC) domains in the C-terminal tail (Fig. 2A).
Figure 2.
Highly correlated regions between Ub and MERS-CoV PLpro. (A) Four highly correlated regions at the contact interface of Ub (stick model) and PLpro (surface model) are shown: FBI (pale green), HCS1 (pale purple), AHC (pale yellow) and UCC (pale orange). (B) The side-chain dihedral angle correlation network showing how mutation leads to conformational changes in distal regions (see Methods for details). The specific dihedral angles are indicated as . (C) Residues targeted for mutation based on interaction networks: R42, A46, K48, E64, V70, R74, and G75. Note that R42 and V70 were not considered in further Ub variant (UbV) designs and experiments after computational prediction.
First, we performed several 500-ns atomic MD simulations to model full protein flexibility of the wtUb–PLpro complex. We employed two principle techniques to study correlated protein motions, i.e., side-chain dihedral angle correlation (Fig. 2B) and force distribution analysis (FDA)36,37 (Fig. S1). Next, we selected highly interactive residues displaying strong correlation in the residue network and also in the Ub–PLpro interface as potential mutation targets in each region (Fig. 2B and C). Notably, previous studies have shown that altering residues near the C-terminus of Ub hampers its biological activity,38–40 so we hypothesized that mutating any residues in the UCC could prevent substrate ubiquitination. MERS PLpro favors binding and cleaving the K48- and K63-linked Ub chains,18,23 which implies that residues near the conjugation points interact frequently with PLpro. Notably, K48 and K63 lie in the HCS1 and FBI domains, respectively. Mutating residues within interactive hotspots may easily hamper PPI, but also offers the potential to strengthen them. Since the FBI and UCC are rather flexible,24 we began by modifying residues A46 and K48 in HCS1 and V70 and R42 in the AHC.
Selection of candidate residues for mutational analysis
Residues A46 and K48 in HCS1 exhibit suboptimal interactions, with weak attraction to surrounding PLpro residues. Specifically, A46 is flanked by Y208 and Y223. To maximize the hydrophobic effect and local nonpolar attraction, we selected a bulkier non-polar amino acid, PHE, for A46 substitution. TYR and TRP were excluded as substitute residues due to spatial constraints. Residue K48 is surrounded by the side-chain of K204. Both K48 and K204 are positively charged, so they are not favored to interact. Accordingly, we selected E48, L48, S48, and I48 as mutation candidates. These substitutions introduce negative, nonpolar, or polar properties, thereby augmenting the potential for enhanced interactions with K204 and Y208 (Fig. 3B). V70 is subjected to weak repulsion from neighboring residues, so the E70 substitution represented a promising mutation due to the ability of its longer and polar side-chain to enhance electrostatic interaction between the G247 backbone and G248 side-chain (Fig. S2).
Figure 3.
Selection of candidate residues suitable for mutational analysis to strengthen localized attraction. (A) Residues targeted for mutation based on interaction networks: R42, A46, K48, E64, V70, R74, and G75. (B) wtUb displays weak interaction with surrounding residues. The A46F and K48E mutations have the potential to increase both vdW and electrostatic interactions. (C) The E64Y mutation increases vdW attraction for G228 and G229. (D) The R74N mutation enhances electrostatic attraction for V275, whereas the G75S mutation increases the attraction for G276. Inward-pointing arrows indicate attraction, outward-pointing arrows represent repulsion. Color-coded regions: Ubiquitin (Ub, orange stick model), PLpro (blue surface model), FBI (pale green), HCS1 (pale blue), AHC (pale yellow), and UCC (pale orange).
In the AHC, R42 presents a strong compensatory effect through strong repulsive forces with the D164 side-chain and an attractive force with the S165 backbone. We generated a D42 mutant variant displaying a negative charge and a shorter side-chain with a view to minimizing the repulsive forces that mainly arise from crossing van der Waal (vdW) radii (Fig. S2).
In the FBI, our dihedral angle correlation network selected E64 and S65 as targets in different MD simulation runs because of the inherent flexibility of the FBI. MERS PLpro unambiguously recognizes K63-linked Ub chains, so we postulated that modifying E64, i.e., between K63 and S65, may simultaneously enhance binding strength and specificity. E64 lies in close proximity to G208 and G209, both of which are nonpolar (Fig. 3C). To maximize nonpolar interactions, we generated 64Y and 64F substitution variants that have large hydrophobic rings.
We targeted R74 and G75 in the UCC for mutation, both of which lie in the LXGG recognition site. Mutating residues of the LXGG recognition site can block the proteolytic cleavage activity of PLpro.24 We tested several dual mutations proposed previously,32 including R74P/R74N and G75R/G75S. From a structural point of view, steric hindrance within the UCC introduces strong vdW repulsion. Therefore, we assumed that the smaller and polar side-chain of the N74 substitution would minimize this repulsive force while retaining polar attraction. We also selected P74 mutation because it could generate localized steric hindrance to stabilize rotation of the surrounding side-chain. R75 and S75 are both polar residues and have the potential to increase local electrostatic interaction relative to nonpolar G75 (Fig. 3D).
To predict the intermolecular attractions between each of our UbV-PLpro variants, we used MM/PBSA to evaluate PPI energy (Fig. S3) and local structural analysis to investigate localized attraction (Fig. S2). Variants harboring the A46 and K48 mutations displayed enhanced UbV–PLpro attraction. However, the K48E-V70E variant did not yield good vdW interactions, and the R42D mutation that resulted in an opposing charge yielded poor predicted UbV–PLpro interaction energies (Table S1 and Fig. S2). These results indicated that mutating R42 or V70 may not promote tight binding, so thereafter we focused on altering the A46 and K48 residues. Additionally, for every designed UbV exhibiting UbV–PLpro attractions that were predicted to be stronger than that of wtUb (Table S1), we also performed PPI-GaMD in an explicit solvent model (to account for solvent effects) to examine their binding residence time41 (Fig. S4). Since PPI-GaMD may enhance conformational sampling, the residence time cannot be compared directly to binding affinity or dissociation rate constant data. Nevertheless, the dissociation times for these UbVs were invariably longer than that determined for wtUb, supporting that these variants likely exhibit tighter binding than wtUb.
Experimental characterization of inhibitory UbVs
Next, we evaluated the UbV-dependent inhibitory efficiencies for MERS PLpro by means of sensitive fluorescent polarization (FP) to monitor the dynamics of fluorescein conjugated to the C-terminus of ISG15 (Fig. 4A). We assessed four designed UbVs harboring the dual mutations of A46F and K48E/S/I/L, as well as the two single-mutation UbVs, i.e., A46F and K48E. These UbVs all efficiently inhibited PLpro enzyme function, displaying reduced IC50 values (Fig. 5B, Table 1 and Fig. S6), and all bound tightly to PLpro (KD values shown in Table 1 and Fig. S7). Relative to wtUb, substitution of A46 in the HCS1 by non-polar PHE resulted in stronger vdW attractions with nearby PLpro residues (Fig. 6A and Table S1). More specifically, A46F induced local conformational arrangements to prompt Y208 and Y223 in PLpro into forming interactions and R233 in PLpro into forming -cation attraction with the UbV (Fig. 6A). Moreover, A46F substitution substantially stabilized overall Ub-PLpro interactions, increasing the KD value 15-fold compared to wtUb (Table S1). The polarity of K48 in wtUb significantly affects both local interactions and network correlation due to the repulsive force with the nearby K204 residue of PLpro. We postulated that mutating K48 to a nonpolar or negatively charged residue such as LEU or GLU would enhance the attractive force between K48 and K204 (Fig. 6A). Notably, the single-point A46F or K48E mutations alone resulted in IC50 values of 1.6 and 3.9 μM, respectively (52.9 μM for wtUb). The respective dual mutant variant further elevated the IC50 value to ~0.2 μM (Table 1), i.e., approximately 250-fold greater PLpro inhibition compared to wtUb. Overall, we observed a synergistic inhibitory effect and enhanced binding affinity for A46F and K48E (or K48L/K48S/K48I) substitutions in terms of PLpro and UbV interactions (Fig. 3C, 4A and Fig. S7).
Figure 4.
The inhibitory IC50 values for designed UbVs. The schematics to the right in (A) represent the cleavage reaction for ISG15* substrate, where * stands for the AVTRYVDC sequence (part of SARS-CoV-2 NSP2) crosslinked with a fluorescein probe. When an UbV is added to the mixture, the substrate binding site is blocked and cleavage is retarded. The grayscale gradient reflects inhibition strength. (B) Monitoring the real-time cleavage reaction by detecting fluorescence polarization. Relative inhibitory activity (IC50 curves) of seven selected UbVs and wtUb reveal how PLpro activity is progressively inhibited as the number of mutations increases from two to five. (C) Experimentally measured IC50 values for 26 UbVs summarized on a logarithmic scale, with black, green, orange, and blue circles indicating weak, medium, strong and, very strong inhibitors, respectively.
Figure 5.
(A, B) BLI sensorgrams (left) and fitting curves (right) for UbV2 and UbV4 show KD values of 0.22 μM and 0.15 μM, respectively. (C) UbV3 and UbV5 bind tightly to PLpro, showing KD values of 2.77 and 1.48 nM, respectively. The fitting values for kon and koff are colored red in the titrated BLI sensorgrams. (D) The IC50 and KD values we measured are strongly correlated (R2 = 0.94). The KD values of ME.2 and ME.4 were published previously.32
Figure 6.
Conformational dynamics and interactions of wtUb and UbVs residues with the HCS1 and FBI regions of the Ub-PLpro complex. Ub and PLpro are shown in orange or blue, respectively. (A) The new attraction forces introduced by the A46F and K48E mutations in the HCS1 region are shown in the right panels. (B) Superimposition of 50 PLpro conformations from a 500-ns MD run. The FBI region (pale green) of the Ub-PLpro complex is highly stabilized by the E64Y mutation, with the hinging motion of backbone residue E230 (pink) in the interaction network contributing significantly to the loop conformations. Wild-type E64 or mutated Y64 of Ub (orange) is shown in stick format.
Above, we have demonstrated that mutating two residues in HCS1 represents an effective strategy for designing binding inhibitors, so we explored mutating additional residues based on local structural analysis. We focused on further modifying the A46F-K48E variant (termed UbV2 hereafter), which exhibited favorable IC50 and KD values. First, we subjected UbV2 to E64Y mutation, representing a residue in the FBI. Experimental measurements of the A46F-K48E-E64Y variant (hereafter denoted UbV3) revealed remarkable binding specificity to and inhibition of PLpro, with an IC50 of 15 nM, i.e., in significant agreement with the strongest computed binding energy (Table 1). Both the root mean square fluctuation (RMSF) and dihedral entropy of PLpro (Fig. S5) were reduced upon interaction with UbV3 compared to UbV2, with the FBI of the UbV3–PLpro complex being more stabilized (Fig. 6B). We measured a KD of 2.77 nM for UbV3-PLpro, which is ~80-fold greater than that determined for UbV2-PLpro. Furthermore, the IC50 value of A46F-E64Y (34 nM) is 48-fold or 8-fold greater than that of A46F or E64Y alone, respectively. Thus, our inhibition assays and binding affinity data greatly support that the three residues at positions 46, 48, and 64 cooperatively stabilize the interaction of Ub with PLpro, hence UbV3 is a better inhibitor of PLpro than UbV2 (Fig. 4C, 5). Altering the surface charge (K48E and E64Y) or hydrophobicity (A46F and E64Y) of the small 76-residue Ub peptide may be structurally destabilizing. Therefore, we employed circular dichroism (CD) spectroscopy to measure the far-UV CD spectra of Ub and nine selected UbVs at 25 °C and 80 °C to validate their thermostability. This analysis revealed that UbV3 displays higher thermostability than the single (A46F, K48E, E64Y) or double-point (UbV2) mutants (Fig. S8).
Ub C-terminal mutations R74N and G75S further stabilize PPIs
Next, we altered two additional residues of UbV2, R74 and G75 of the UCC region based on information from our non-covalent correlation network, with these residues serving a role in preventing cells from recognizing the UbVs. The four-point A46F-K48E-R74N-G75S mutant variant (hereafter termed UbV4) yielded a reduced IC50 of 110 nM. Similar to the data for double mutants, the K48E/K48L/K48S/K48I mutations resulted in the same inhibitory effect (IC50: 110–290 nM, Table S1). Therefore, we further included the E64Y substitution in UbV4 to encompass all three highly correlated regions, with the resulting designed five-point UbV5 mutant displaying an IC50 value of 9.7 nM (i.e., 5,500-fold that of wtUb) and a KD of 1.5 nM (Table 1). The characterized KD values for UbV3 and UbV5 are approximately 20-fold greater than those established for the phage-display-screened UbVs ME.2 and ME.4 generated in a previous study32 (Table S1). In addition to their impressive attribute of functional PLpro inhibition, both UbV3 and UbV5 bound highly specifically to MERS PLpro (Fig. S9), which is also an essential attribute for a good inhibitor. Thus, our UbV3 and UbV5 variants exhibit strong inhibition, equivalent to reported variants for MERS PLpro.32 In contrast to the preserved thermostability of the UbVs designed in this study, the previously reported ME.2 and ME.4 variants harboring 15 mutated residues denatured at 80 °C,33 and thus are significantly less thermostable than our UbVs (Fig. S8). Similarly, the Tm value of the nine-point U7Ub25.2540 mutant for USP7 is 64 °C lower than that of wtUb.33 Consequently, mutating too many residues in Ub can easily elicit stability issues. Our study demonstrates the advantage and efficacy of modifying only a few residues of a protein template to enhance PPI.
In this study, we established a novel approach to computationally select key residues responsible for PPI based on dihedral angle networks. We found that the mapped dihedral angle networks are useful for identifying critical interactions between proteins. Then, we used this information to select residues for mutation to alter PPI strengths. Using Ub-PLpro as our model system, we have demonstrated that modifying only two or three residues within the correlation network of the Ub–PLpro interface successfully enhanced PPI and resulted in a 250- to 3,500-fold reduction of MERS PLpro activity. A combination of five mutated residues in the HCS1, FBI and UCC domains of the Ub-PLpro complex resulted in a 5,500-fold (IC50 = 9.7 nM) reduction in PLpro activity and a 27,500-fold enhancement in UbV–PLpro complex affinity. Our design platform can be used to computationally examine a correlation network of protein side-chains and local pair-wise forces to efficiently design UbVs for further experimental assessment. The considerable correlation between IC50 and KD values (R2 = 0.94) implies that our experimental IC50 data can be used to indirectly estimate KD values for UbV-PLpro complexes. Integrating experimental measurements and structural analyses using MD simulations, together with appropriate post-hoc analysis, can iteratively inform new designs.
Our strategy is transformative and highly efficient for identifying key mutation sites and specific residues to guide rational design of many disease-linked DUBs and Ub-bound proteins,30,31 including USP4,42 USP7,43 USP11,44 and PLpro of SARS-CoV-2.30 Apart from producing variant-specific anti-bodies/vaccines for diverse spike proteins of viruses, blocking the functions of viral nonstructural proteins represents an alternative therapeutic solution to tackle COVID-19 and other coronaviruses. Designing tightly binding and specific UbV inhibitors for the coronavirus PLpro complex is an elegant solution to retard viral replication and simultaneously rescue the host antiviral immune response. This same strategy can be applied straightforwardly to other protein–protein interacting systems related to signaling and enzymatic activity. As these cellular events frequently reoccur, the protein complexes involved in cell signaling and enzymatic reactions do not have perfectly optimized PPI as stably bound complexes. Therefore, mutating residues in their interacting regions can remarkably improve binding. Overall, modifying residues displaying highly correlated dynamic motion can be used to engineer tightly binding variants for various applications.
Materials and Methods
MD simulation protocol
The X-ray crystal structure of MERS-CoV-PLpro-wtUb was obtained from the RCSB protein data bank (PDBID: 4RF0).26 PLpro was extended by one residue at the N-terminus, and wtUb was extended by two residues to ensure consistent numbers of residues for comparison with preexisting UbVs such as ME.2.32 PLpro comprises 319 residues and Ub comprises 78 residues. The Molecular Operating Environment platform45 was used to perform all UbV mutations. All MD simulations were performed in the AMBER 20 package with GPU acceleration46 Force Field ff14sb47 was applied to proteins. First, we minimized the hydrogen atoms, amino acid side-chain, and the entire protein system for 500, 1000, and 5000 steps, respectively, in a generalized Born implicit solvent. All systems were then solvated in TIP3P water, with an extension of 12 Å from the solute edge. Two Cl- counter-ions were added to neutralize the charge of the system. The solvated system contains approximately 72,000 atoms. The water molecules were minimized for 1000 steps, followed by minimization of the entire system for 2000 steps. The solvated system was equilibrated under constant pressure and temperature (NPT ensemble) from 50 K to 275 K with 25 K increments and 100 ps each, and finally at 298 K for 500 ps. Production runs were also performed in the NPT ensemble at 298 K using a Langevin Thermostat with 2-fs time-steps. The first 50 ns of the MD simulation were treated as equilibrium plus. Force Distribution Analysis (FDA)36,37 and molecular mechanics Poisson-Boltzmann surface area (MM/PBSA) calculations were performed using data from the subsequent 450 ns. The cutoff for nonbonding interactions, which includes vdW and electrostatic components, was set to 12 Å. The Particle Mesh Ewald Method was used to compute long-range electrostatic interactions.
It is possible that a large protein–protein system becomes trapped at a specific local minimum, resulting in suboptimal outcomes. We performed three independent 150-ns MD simulations for each conformation and selected the lowest energy trajectories by calculating MM/PBSA energies. The exterior dielectric constant was set to 15 to accommodate the polar protein residues at the protein-water interface. The trajectories with the lowest energy were extended to a 500-ns production run. Output trajectories were saved every 1 ps for further analysis.
Side-chain dihedral angle correlation network
The following steps were used as a rationale for selecting residues for substitution:
1. Construct a correlation network of side-chain dihedral angles for wtUb-PLpro
We used T-analyst48 to calculate the side-chain dihedral rotations of each amino acid residue and their pairwise correlations. Each side-chain dihedral angle was recorded every 100 ps through 500 ns trajectories to generate 5000 different angles per dihedral selection. Pairwise correlations were computed using a Pearson correlation formula. We converted the side-chain dihedral angles to Cartesian coordinates by means of equations (1)-(3) to accurately capture their differences and means, thereby preventing erroneous computation of their correlation at the discontinuity margin (±180° or 360°/0°).48 Notably, positive correlation between two side-chains indicates that the two sides rotate similarly during MD simulation.
| (1) |
| (2) |
| (3) |
From the pairwise correlation matrix, we generated a correlation network by using python library NetworkX,49 enabling us to visualize the correlation between each residue. Specific side-chain rotation can generate a chain effect and impact the rotation of distal residues. We applied a correlation cutoff of 0.3 to eliminate less correlated residues. The Ub residues selected for further analysis were: I3, T14, T22, E24, K27, I30, E34, R42, K48, Q62, S65, T66, H68, V70, L71, L73, R74, and D77.
2. Define the contact interface and eliminate residues not at the contact interfaces
We adopted FDA to select the contact interface between wtUb and PLpro. A cutoff of 10 (pN) was applied to disregard areas of low interaction. Next, we generated a heatmap of pairwise forces between wtUb and PLpro (Fig. S1). From this heatmap, we identified four clear interface regions: HCS1, AHC, FBI, and UCC. Some highly correlated residues selected from the previous step are not located at the contact interface. Residues located outside the contact interface contribute little or no interaction to binding affinity, so they were discarded from further analysis. Ub residues selected for analysis: R42, K48, Q62, S65, V70, L71, L73, and R74.
3. Consider multiple residues at the FBI
The FBI region is highly dynamic, displaying high RMSF values and it has been shown previously to crystallize in different conformations24 Our FDA data also revealed differential patterns among randomly-seeded production runs. Notably, our dihedral angle correlation network selected residues S65 and T66, as well as K63 on the conjugated side of PLpro. We postulated that mutating E64, which lies between S65 and K63, could further optimize our results. Ub residues selected for analysis based on this step: R42, K48, Q62, E64, S65, T66 H68, V70, L71, L73, and R74.
4. Include ALA, GLY and PRO residues
ALA, GLY and PRO residues do not have side-chains, so they would have been excluded from dihedral angle selection. To avoid excluding potentially useful mutation sites, we included all ALA, GLY and PRO residues within 5 Å of our previously selected mutation sites. Ub residues selected for analysis from this step: R42, A46, K48, Q62, E64, S65, T66, H68, V70, L71, L73, R74, and G75.
5. Further select residues based on FDA data
Residues of interest displaying strong attraction to their surrounding residues will not have been selected for mutation in the previous steps. Accordingly, we sought residues that displayed strong repulsion or weak interactions with their surrounding residues. A46 interacts only weakly with its surrounding residues, and K48 has weak interactions with surrounding residues K204, Y208, and V209 of PLpro at the HCS1. Therefore, we felt that A46 and K48 represented ideal residues for mutation. In addition, R42, V70, and G75 presented repulsive forces with their surroundings and accordingly were chosen for mutation. In contrast, L71 and L73 in the UCC exert strong attractive forces on the BL2 and AHC domains, so they were excluded from mutational analysis. Although E64 and R74 present strong attractive forces to their surroundings, we still considered them as possible mutational sites because both the dihedral angle correlation network and FDA results from different random seeds resulted in large standard deviations at the FBI and UCC because of their highly flexible nature (Fig. S1). Moreover, R74 is part of the LRGG recognition site, with mutation of R74 further hindering proteolytic cleavage.24 Accordingly, only seven residues were ultimately selected for mutational analysis: R42, A46, K48, E64, V70, R74, and G75.
PPI-GAMD simulation protocol
Starting from the last frame of our MD simulation, we performed 5 ns of classical MD simulation, followed by 5 ns of PPI-GAMD equilibration to obtain the boost parameters (ntcmdprep = 500,00 0, ntcmd = 2,500,000, ntebprep = 500,000, and nteb = 2,500,000 steps). Production runs were performed in the NPT ensemble at 350 K using a Langevin Thermostat with a 2-fs time-step. We applied both potential boost and dihedral boost (igamd = 17) on Ub residues within 5 Å of the PLpro residues at the contact interfaces. Applying dual boost potentials on the entire Ub structure can result in denaturation of the protein structure, leading to suboptimal results. The threshold energy of potential boost was set to the upper bound limit (iEP = 2), and the threshold energy for dihedral boost was set to the lower bound limit (iED = 1). The upper limit of the standard deviation for dual boost potential was set to 10 kcal/mol (sigma0P and sigma0D = 10). The production run continued until we observed Ub dissociating from PLpro. We defined dissociation according to a sudden increase in Cα RMSD values. Production runs were repeated with three different random seeds, with the longest dissociation times being reported herein.
Protein expression and purification
Genes encoding SARS-CoV-2 PLpro, MERS PLpro, ISG15 and ubiquitin were synthesized by GenScript (NJ, USA). Ubiquitin variants were made by site-directed mutagenesis or directed amplification (C-terminal mutations). UbV genes ME.2 and ME.4 were synthesized and subcloned by Genomics (Genomics Inc., Taiwan). All genes were placed in pRSFDuet-1 vector with a N-terminal hexahistidine tag (his-tag) and a TEV cleavage sequence. All resulting plasmids were transformed into the BL21 RIL cell line for protein production. For PLpro and ISG15-AVTRYVDC, E. coli grown in LB medium at 37 °C to an OD600 of 0.6–0.8 was induced by treatment with 0.6 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) overnight at 16 °C (MERS PLpro and SARS-CoV-2 PLpro) or at 25 °C (ISG15-AVTRYVDC). For all 26 UbVs assessed in the current study, E. coli was grown in autoinduction medium containing base broth (25 mM Na2HPO4, 25 mM KH2PO4 pH 7.2, 85 mM NaCl, 0.5% yeast extract, 2% tryptone) and a sugar mix (15% v/v glycerol, 1.25% w/v glucose, 5% w/v lactose) at a 25:1 volume ratio. The E. coli was cultured at 37 °C for 24 hours, with UbV proteins being automatically expressed once glucose had been depleted.
E. coli cell pellets were spun down and resuspended in Buffer A (25 mM Tris-HCl pH 7.6, 200 mM NaCl, 3 mM 2-Mercaptoethanol) with the addition of 1 mM PMSF for sonication. Cell lysates were then centrifuged, before loading the supernatant on Roche cOmplete nickel resin and then washing and eluting it using 300 mM imidazole. The his-tag of PLpro was removed by means of TEV protease treatment, and the resulting proteins were further purified by size exclusion chromatography (SEC) on an Akta FPLC system (Cytiva). ISG15-AVTRYVDC was further crosslinked with fluorescein-5-Maleimide (Santa Cruz Biotechnology) at 4 °C for 1 hour. Excess fluorescein was removed by processing through desalting columns. All proteins were flash-frozen in liquid nitrogen, aliquoted and stored at —80 °C.
Fluorescence polarization (FP)-based inhibition assay
To detect and characterize the inhibitory effects on PLpro by UbVs, we used fluorescein-labeled ISG15-AVTRYVDC (denoted ISG15* hereafter), where AVTRYVD is the N-terminal sequence of SARS-CoV-2 and a CYS residue was introduced at the C-terminal end for crosslinking with the Fluorescein-5-maleimide (Ana Spec). Values of fluorescence polarization (FP) for ISG15* and the cleaved AVTRYVDC* are approximately 120 and 20, respectively (Fig. S6), representing a sensitive tool to unravel the activity of MERS PLpro. We measured 40-μl samples in multiple wells composed of 2 μM ISG15*, 50 nM MERS PLpro (or SARS-CoV-2 PLpro), and a wide range of UbV concentrations (i.e., from 0.06 nM – 150 μM) in a 384-well plate for 1,800 seconds or longer. ISG15* alone was used as a control. Assessments of individual UbV concentrations were conducted in triplicate to determine standard errors.
The reaction rate constants (kobs) of ISG15* cleavages were obtained by curve fitting using one-phase decay (equation (4). Enzymatic activity was normalized according to the ratio of the kobs values with or without UbV. To determine IC50 values, normalized UbV concentration-dependent enzymatic activities were fitted according to the logistic non-linear regression model (equation (5).
| (4) |
where is the value when (time) is zero, is a plateau value at infinite time, and is the rate reaction constant.
| (5) |
The values (representing the FP readouts) were normalized to between 0 and 1.0 for the fitting that included “Hill slope”, with all of the values being ~1.0.
Bio-Layer interferometry (BLI)
BLI experiments were performed on an Octet RED 96 system (Sartorius) using anti-GST antibody biosensors for GST-tagged MERS PLpro and UbV (or Ub) as analytes at 25 °C. The ligand and analytes were diluted into reaction buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1 mg/ml BSA, 0.01% Tween-20). Steady-state response wavelength shifts of analytes in multiple concentrations were used to fit a single-site binding system and determine the dissociation constant according to equation (6).
| (6) |
where is the steady-state response shift of the sensorgram curve, [C] is the concentration of analytes, is the maximal response, and is the dissociation constant. To determine and values, we applied a Levenberg–Marquardt algorithm to perform iterative non-linear least squares curve fitting.
The and values of UbV3 and UbV5 were globally fitted to the time-dependent response wavelength shifts in the association and dissociation well, respectively, using the Octet Data Analysis software (Sartorius). values for UbV3 and UbV5 were calculated as .
Circular dichroism (CD) spectroscopy
CD measurements were performed on a Jasco J-815 spectropolarimeter. Far-UV spectra were measured from 260 to 195 nm at 25 and 80 °C. UbV samples (10 μM) diluted in 25 mM Tris pH 7.6 and 50 mM NaCl buffer were measured in a 1 mm quartz cell (Hellma GmbH). Melting temperature experiments were performed at 25 and 80 °C at 1 °C intervals. The CD spectra were averaged from triplicates acquired at a scanning speed of 50 nm/min and a digital integration time of 1 second.
Supplementary Material
Acknowledgments
This study was supported by the US National Institutes of Health (GM-109045), the US National Science Foundation (MCB-1350401 to C.C), and an Academia Sinica (Taipei, Taiwan) Career Development Award (AS-CDA-110-L03 to K.P.W.), and the National Science and Technology Council (110-2311-B-001-046 and 111-2113-M-001-026-MY2 to K.P.W.). We appreciate the technical and instrumental support from the Academia Sinica (AS) Biophysics Core Facility (AS-CFII-111-201) and the Biophysics Instrumentation Laboratory, Institute of Biological Chemistry, Academia Sinica.
Footnotes
Code Availability
Source code for the dihedral angle correlation network analysis is available upon request.
Data and Materials Availability
All data used in the main text or the supplementary materials are available on request.
CRediT authorship contribution statement
Ta I Hung: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Yun-Jung Hsieh: Conceptualization, Methodology, Investigation, Formal analysis, Writing – original draft, Writing – review & editing. Wei-Lin Lu: Methodology, Formal analysis. Kuen-Phon Wu: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition. Chia-en A. Chang: Conceptualization, Methodology, Writing – review & editing, Supervision, Funding acquisition.
DECLARATION OF COMPETING INTEREST
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2023.168337.
DATA AVAILABILITY
Data will be made available on request.
References
- 1.Zahradník J, Schreiber G, (2021). Protein engineering in the design of protein-protein interactions: SARS-CoV-2 inhibitors as a test case. Biochemistry 60, 3429–3435. 10.1021/acs.biochem.1c00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marchand A, Van Hall-Beauvais AK, Correia BE, (2022). Computational design of novel protein–protein interactions – an overview on methodological approaches and applications. Curr. Opin. Struct. Biol. 74, 102370. 10.1016/j.sbi.2022.102370. [DOI] [PubMed] [Google Scholar]
- 3.Petrey D, Zhao H, Trudeau SJ, Murray D, Honig B, (2023). PrePPI: a structure informed proteome-wide database of protein-protein interactions. J. Mol. Biol, 168052. 10.1016/j.jmb.2023.168052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao L, Coventry B, Goreshnik I, Huang B, Sheffler W, Park JS, Jude KM, Markovic I, et al. , (2022). Design of protein-binding proteins from the target structure alone. Nature 605, 551–560. 10.1038/s41586-022-04654-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frappier V, Keating AE, (2021). Data-driven computational protein design. Curr. Opin. Struct. Biol. 69, 63–69. 10.1016/j.sbi.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pearce R, Huang X, Setiawan D, Zhang Y, (2019). EvoDesign: designing protein-protein binding interactions using evolutionary interface profiles in conjunction with an optimized physical energy function. J. Mol. Biol. 431, 2467–2476. 10.1016/j.jmb.2019.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rooklin D, Modell AE, Li H, Berdan V, Arora PS, Zhang Y, (2017). Targeting unoccupied surfaces on protein-protein interfaces. J. Am. Chem. Soc. 139, 15560–15563. 10.1021/jacs.7b05960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pahari S, Li G, Murthy AK, Liang S, Fragoza R, Yu H, Alexov E, (2020). SAAMBE-3D: predicting effect of mutations on protein-protein interactions. Int. J. Mol. Sci. 21, 2563. 10.3390/ijms21072563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kudlacek ST, Metz S, Thiono D, Payne AM, Phan TTN, Tian S, Forsberg LJ, Maguire J, et al. , (2021). Designed, highly expressing, thermostable dengue virus 2 envelope protein dimers elicit quaternary epitope antibodies. Sci. Adv. 7 10.1126/sciadv.abg4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenfeld L, Heyne M, Shifman JM, Papo N, (2016). Protein engineering by combined computational and in vitro evolution approaches. Trends Biochem. Sci. 41, 421–433. 10.1016/j.tibs.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Lite T-L-V, Grant RA, Nocedal I, Littlehale ML, Guo MS, Laub MT, (2020). Uncovering the basis of protein-protein interaction specificity with a combinatorially complete library. Elife 9 10.7554/eLife.60924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parkinson J, Hard R, Wang W, (2023). The RESP AI model accelerates the identification of tight-binding antibodies. Nature Commun. 14, 454. 10.1038/s41467-023-36028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swatek KN, Komander D, (2016). Ubiquitin Modifications. Nature Publishing Group, pp. 399–422 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Komander D, Rape M, (2012). The ubiquitin code. Annu. Rev. Biochem 81, 203–229. 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 15.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP, (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340. 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrigan JA, Jacq X, Martin NM, Jackson SP, (2017). Deubiquitylating enzymes and drug discovery: emerging opportunities. Nature Rev. Drug Discov. 17, 57–78. 10.1038/nrd.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrow JK, Lin HK, Sun SC, Zhang S, (2015). Targeting ubiquitination for cancer therapies. Future Med. Chem. 7, 2333. 10.4155/FMC.15.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Békés M, Rut W, Kasperkiewicz P, Mulder MPC, Ovaa H, Drag M, Lima CD, Huang TT, (2015). SARS hCoV papain-like protease is a unique Lys48 linkage-specific di-distributive deubiquitinating enzyme. Biochem. J 468, 215–226. 10.1042/BJ20141170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey-Elkin BA, Knaap RCM, Kikkert M, Mark BL, (2017). Structure and function of viral deubiquitinating enzymes. J. Mol. Biol. 429, 3441–3470. 10.1016/j.jmb.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harcourt BH, Jukneliene D, Kanjanahaluethai A, Bechill J, Severson KM, Smith CM, Rota PA, Baker SC, (2004). Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 78, 13600–13612. 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, et al. , (2020). Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587, 657–662. 10.1038/s41586-020-2601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, Lu BG, Kuchel NW, et al. , (2020). Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 39 10.15252/embj.2020106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Báez-Santos YM, Mielech AM, Deng X, Baker S, Mesecar AD, (2014). Catalytic function and substrate specificity of the papain-like protease domain of nsp3 from the Middle East respiratory syndrome coronavirus. J. Virol. 88, 12511–12527. 10.1128/JVI.01294-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Báez-Santos YM, St SE, John ADM, (2015). The SARS-coronavirus papain-like protease: structure, function and inhibition by designed antiviral compounds. Antiviral Res. 115, 21–38. 10.1016/j.antiviral.2014.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.United Nations, World Economic Situation and Prospects 2022, New York, 2022. [Google Scholar]
- 26.Bailey-Elkin BA, Knaap RCM, Johnson GG, Dalebout TJ, Ninaber DK, Van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL, (2014). Crystal structure of the middle east respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J. Biol. Chem. 289, 34667. 10.1074/JBC.M114.609644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lei J, Kusov Y, Hilgenfeld R, (2018). Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antiviral Res. 149, 58–74. 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Chen X, Bian G, Tu J, Xing Y, Wang Y, Chen Z, (2014). Proteolytic processing, deubiquitinase and interferon antagonist activities of Middle East respiratory syndrome coronavirus papain-like protease. J. General Virol. 95, 614–626. 10.1099/VIR.0.059014-0/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- 29.Zhang W, Wu K-P, Sartori MA, Kamadurai HB, Ordureau A, Jiang C, Mercredi PY, Murchie R, et al. , (2016). System-wide modulation of HECT E3 ligases with selective ubiquitin variant probes. Mol. Cell 62, 121–136. 10.1016/j.molcel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Vliet VJE, Huynh N, Palà J, Patel A, Singer A, Slater C, Chung J, van Huizen M, et al. , (2022). Ubiquitin variants potently inhibit SARS-CoV-2 PLpro and viral replication via a novel site distal to the protease active site. PLoS Pathog. 18, e1011065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ernst A, Avvakumov G, Tong J, Fan Y, Zhao Y, Alberts P, Persaud A, Walker JR, et al. , (2013). A strategy for modulation of enzymes in the ubiquitin system. Science (1979) 339, 590–595. 10.1126/science.1230161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W, Bailey-Elkin BA, Knaap RCM, Khare B, Dalebout TJ, Johnson GG, van Kasteren PB, McLeish NJ, et al. , (2017). Potent and selective inhibition of pathogenic viruses by engineered ubiquitin variants. PLoS Pathog. 13 10.1371/journal.ppat.1006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Zhou L, Rouge L, Phillips AH, Lam C, Liu P, Sandoval W, Helgason E, et al. , (2012). Conformational stabilization of ubiquitin yields potent and selective inhibitors of USP7. Nature Chem. Biol. 9, 51–58. 10.1038/nchembio.1134. [DOI] [PubMed] [Google Scholar]
- 34.Sun MGF, Seo MH, Nim S, Corbi-Verge C, Kim PM, (2016). Protein engineering by highly parallel screening of computationally designed variants. Sci. Adv. 2 10.1126/SCIADV.1600692/SUPPL_FILE/1600692_TABLE_S3.XLSX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee H, Lei H, Santarsiero BD, Gatuz JL, Cao S, Rice AJ, Patel K, Szypulinski MZ, et al. , (2015). Inhibitor recognition specificity of MERS-CoV papain-like protease may differ from that of SARS-CoV. ACS Chem. Biol. 10, 1456–1465. 10.1021/cb500917m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E, (2015). GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 37.Costescu BI, Gräter F, (2013). Time-resolved force distribution analysis. BMC Biophys. 6, 5. 10.1186/2046-1682-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Duerksen-Hughes PJ, Xu X, Wilkinson KD, (1987). Structure and function of ubiquitin: evidence for differential interactions of arginine-74 with the activating enzyme and the proteases of ATP-dependent proteolysis. Biochemistry 26, 6980–6987. 10.1021/bi00396a019. [DOI] [PubMed] [Google Scholar]
- 39.Burch TJ, Haas AL, (1994). Site-directed mutagenesis of ubiquitin, differential roles for arginine in the interaction with ubiquitin-activating enzyme. Biochemistry 33, 7300–7308. 10.1021/bi00189a035. [DOI] [PubMed] [Google Scholar]
- 40.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L, (2001). Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276, 30483–30489. 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Miao Y, (2022). Protein-protein interaction-gaussian accelerated molecular dynamics (PPI-GaMD): characterization of protein binding thermodynamics and kinetics. J. Chem. Theory Comput. 18, 1275–1285. 10.1021/acs.jctc.1c00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clerici M, Luna-Vargas MPA, Faesen AC, Sixma TK, (2014). The DUSP–Ubl domain of USP4 enhances its catalytic efficiency by promoting ubiquitin exchange. Nature Commun. 5, 5399. 10.1038/ncomms6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kategaya L, Di Lello P, Rouge L, Pastor R, Clark KR, Drummond J, Kleinheinz T, Lin E, et al. , (2017). USP7 small-molecule inhibitors interfere with ubiquitin binding. Nature 550, 534–538. 10.1038/nature24006. [DOI] [PubMed] [Google Scholar]
- 44.Wu H-C, Lin Y-C, Liu C-H, Chung H-C, Wang Y-T, Lin Y-W, Ma H-I, Tu P-H, et al. , (2014). USP11 regulates PML stability to control Notch-induced malignancy in brain tumours. Nature Commun. 5, 3214. 10.1038/ncomms4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chemical Computing Group ULC, Molecular Operating Environment (MOE), (2023). [Google Scholar]
- 46.Case DA Walker Ross C, Roitberg Kenneth Merz Pengfei L AM, Amber 2020 Reference Manual Principal contributors to the current codes, n.d. http://ambermd.org/contributors.html. [Google Scholar]
- 47.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C, (2015). ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theory Comput. 11, 3696–3713. 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ai R, Qaiser Fatmi M, Chang CA, (2010). T-Analyst: a program for efficient analysis of protein conformational changes by torsion angles. J. Comput. Aided Mol. Des. 24, 819–827. 10.1007/s10822-010-9376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagberg AA, Schult DA & Swart PJ (2008). Exploring Network Structure, Dynamics, and Function Using NetworkX. http://conference.scipy.org/proceedings/SciPy2008/paper_2 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.