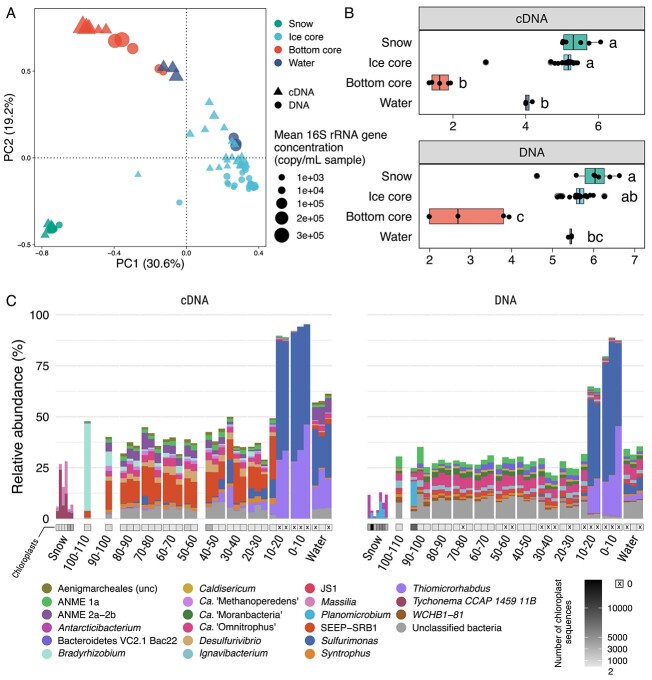
Figure 1.
(A) Principal component analysis showing bacterial and archaeal beta diversity in the Lagoon Pingo ice-covered system. Colors show the sample’s environment (snow, water, ice), and the bottom core was defined based on the dominance of Sulfurimonas. The size of the points show the bacterial 16S rRNA gene concentration in copy number/mL of sample, as determined by quantitative polymerase chain reaction. (B) Shannon index for the four environments presented in (A). Letters indicate groups of significance (False Discovery Rate-corrected P value <.05) representing Dunn’s test output after Kruskal-Wallis significance tests. Observed ASV numbers are available in Supplementary Fig. 4. (C) Genus affiliation of the 20 most abundant ASVs in the cDNA (left) and DNA dataset (right). The x axis shows the height of the ice core (in cm) relative to the water–ice interface (0 cm). Shades of grey below the bars indicate the number of chloroplast reads identified in the sample; a cross on white background indicates the absence of chloroplast read. ASV, amplicon sequence variant.