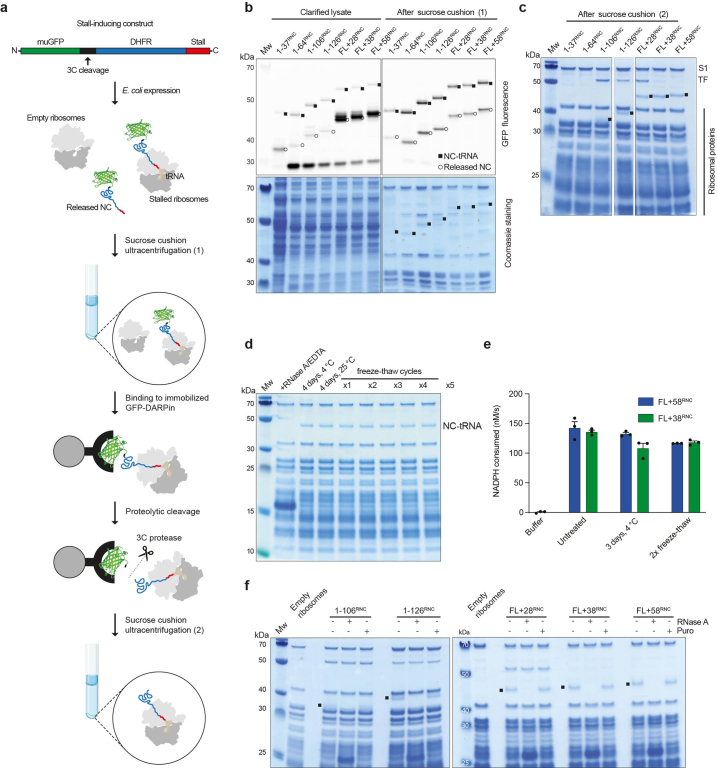
Extended Data Fig. 1. Preparation and quality control of stalled ribosome:nascent chain complexes (RNCs).
a, RNC purification. Constructs encode an N-terminal muGFP followed by a cleavage site for 3 C protease, DHFR sequence, and a C-terminal stalling sequence derived from M. succiniciproducens SecM. Expression in E. coli produces both stalled RNCs and free NCs. Total ribosomes are pelleted by sucrose cushion centrifugation, then RNCs are purified using a GFP-binding DARPin. RNCs are selectively eluted using 3 C protease, leaving GFP on the resin. A second ultracentrifugation step removes residual free NC. b, RNCs are tracked by SDS-PAGE followed by either fluorescent imaging of the GFP or Coomassie staining. Ribosome-bound NCs are covalently coupled to peptidyl tRNA ( ~ 20 kDa) and can therefore be distinguished from released NCs. Note that shortest RNC (1-37RNC) contains an additional disordered linker after the GFP to improve capture by the DARPin. It therefore migrates slower on SDS-PAGE than 1-64RNC. Residual released NC, detectable by sensitive fluorescence imaging, is removed by the second round of centrifugation. Experiments were repeated 5 times with similar results. c, SDS-PAGE with Coomassie staining of purified RNCs. Ribosomal proteins including S1 are indicated, as is Trigger factor. Where visible, the NC-tRNA band is indicated. Experiments were repeated 5 times with similar results. d, RNCs are stable over prolonged incubation and multiple freeze-thaw cycles. The stability of FL + 58RNC was monitored by the integrity of the NC-tRNA band on SDS-PAGE. Conditions included incubation for 4 days at 25 °C or 4 °C, or repeated cycles of freezing in liquid N2 followed by thawing on ice. As a positive control, the RNC was disrupted by treatment with 50 µg/mL RNaseA and 50 mM EDTA. e, DHFR in full-length RNCs is stable over prolonged incubation and multiple freeze-thaw cycles. Enzyme activity of FL + 38RNC and FL + 58RNC was measured either immediately after purification, after incubation at 4 °C for 3 days, or after two freeze-thaw cycles as described in d. Data are presented as mean values ± SD, n = 3 independent experiments. f, Purified RNCs were treated with either 2.5 mM puromycin or 50 µg/mL RNaseA. The NC-tRNA band is indicated in the untreated control lanes. Experiments were repeated twice with similar results.