Abstract
Introduction
Knowledge about the distribution and frequency of the respective haplotypes on the wildtype and mutant allele is highly relevant in the context of future gene therapy clinical studies in Spinocerebellar Ataxia Type 3, the most common autosomal dominantly inherited ataxia. Single nucleotide polymorphisms associated to the disease-causing gene, ATXN3, have been determined. We wanted to investigate the frequency and regional distribution of two intragenic single nucleotide polymorphisms (SNPs) in a large European SCA3 cohort and their relation to the clinical phenotype.
Methods
The genotypes of the two polymorphisms at base pair positions 987 and 1118 of the ATXN3 were determined for their co-localization on the normal and expanded allele, respectively, in 286 SCA3 mutation carriers and 117 healthy controls from 11 European sites.
Results
The distribution of genotypes on the expanded allele differed from those of the wildtype allele of SCA3 mutation carriers and of healthy controls, and was mainly influenced by the regional origin. In our cohort, no particular clinical phenotype was associated with any specific haplotype.
Conclusions
Our results confirm distinct allocations of SNPs associated to the expanded ATXN3, and accordingly the consideration of allele-specific therapies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00415-024-12829-9.
Keywords: SCA3, Polymorphism, ATXN3, Spinocerebellar ataxia, SNP, ASO
Introduction
Spinocerebellar ataxia type 3, SCA3, also known as Machado-Joseph Disease, MJD, is caused by a coding elongated CAG repeat expansion and is one of the most common autosomal dominantly inherited ataxias [1]. SCA3 has a clinical onset in adulthood and is characterized by progressive ataxia, which can be accompanied by additional neurological symptoms such as oculomotor dysfunction or spasticity [2]. The highest prevalence is found in the Azores (Portugal) [1, 3].
The disease-determining CAG repeat expansion is located on chromosome 14q32.12, in the protein coding region of the ATXN3 gene [4–6]. Wildtype alleles usually have a length of up to 44 CAG repeats [1, 7, 8]. Due to the CAG repetition on the expanded allele, with around 60–87 repeats, the resulting ATXN3 protein exhibits an expanded polyglutamine (polyQ) tract causing dysfunction of the altered protein [1, 9]. Such mutant ATXN3 is prone to aggregation and responsible for a “toxic gain of function” disturbing cellular homeostasis [10, 11]. Currently, there is no disease-modifying treatment available and the clinical management remains symptomatic and supportive. However, first clinical trials with gene therapy approaches such as antisense oligonucleotides (ASO) have been initiated (https://clinicaltrials.gov, NCT05160558, NCT05822908). ASOs and other approaches including small interfering RNAs (siRNA) aiming at silencing the ATXN3 gene are currently promising therapeutic options in SCA3 [12–17]. Importantly, a general aim of gene therapies is to reduce the disease protein in an allele-specific manner to preserve the physiological function of the wildtype allele. Consequently, single nucleotide polymorphisms (SNPs) that are linked to the ATXN3 gene, especially to the expanded allele, are of particular interest for an allele-specific gene therapy approach. The knowledge about the frequency and regional distribution of the related haplotypes is of central relevance regarding personalized therapies.
Several SNPs associated to the ATXN3 gene have been described particularly with regard to the expanded, disease-determining allele [18–21]. Two well-described SNPs showed an association with the disease determining allele: first, rs12895357 at base-pair (bp) 916 (c.916G > C; p.Gly306Arg) is historically known and thus also in the present work labeled as bp987, and second, rs7158733 at bp1118 (c.1118C > A; p.Tyr349*) [4, 21–23]. These two SNPs are part of the haplotype described in the two major ancestral origins of SCA3: (i) the Machado lineage, determined by the GTGGCA haplotype, which is geographically more restricted to the Azorean island of São Miguel and mainland Portugal, and (ii) the TTACAC haplotype, also known as the Joseph lineage, is found in the ancient mutation origin in Asia, which later spread throughout Europe, especially on the Portuguese mainland and on the Azorean island of Flores [21, 24]. Several studies showed the segregation of the C-A haplotype on the expanded allele in the majority of the families of various ethnic backgrounds including Portugal (Azores and mainland), Brazil, Spain, Taiwan, Germany, Japan, France, UK, India, US, Cambodia, and China [4, 21, 22, 25].
The determination of SNPs is not part of standard diagnostic genetic testing and characterizations of SNPs in large cohorts of SCA3 mutation carriers are up to now missing. In particular, the frequency and regional distribution of the two SNPs linked to the expanded allele across Europe remains elusive, even though allele-specific protein-lowering therapies covering bp987 and bp1118 are already under development [12, 26]. The aim of this work was to study the frequency and regional distribution of the two above-mentioned SNPs (bp987 and bp1118) in a large European cohort of 280 SCA3 mutation carriers and to determine the relation of the intragenic SNPs to the clinical phenotype.
Methods
Participants
All participants were included in the longitudinal observational study European Spinocerebellar Ataxia Type 3/Machado-Joseph-Disease Initiative (ESMI) and gave their written informed consent according to the declaration of Helsinki. The inclusion criteria were as follows: (i) age ≥ 18 years, (ii) known SCA3 mutation carrier, or first-degree relative of a SCA3 mutation carrier or healthy control. Participants were recruited at 11 European research centers (London, UK; Bonn, Aachen, Essen, Tübingen and Heidelberg, Germany; Coimbra and the Azores, Portugal; Nijmegen and Groningen, The Netherlands; Santander, Spain). EDTA blood samples were taken following a standardized protocol [27]. In total, DNA samples from 286 SCA3 mutation carriers and 117 healthy controls were genotyped for the SNPs rs12895357 at bp987 and rs7158733 at bp1118 of the ATXN3 gene. As demographic and genetic data, age at baseline and sex as well as the CAG repeat length of both alleles were extracted. Determination of CAG repeat length was assessed centrally for all participants at the Institute of Medical Genetics and Applied Genomics, Tübingen.
Two SCA3 mutation carriers were homozygous for the disease-specific mutation. They were excluded from the main analysis to maintain statistical conformity. Their descriptive information is provided in a separate paragraph of the results’ section.
Clinical assessment included the Scale for the Assessment and Rating of Ataxia (SARA) [28] and the Inventory of Non-Ataxia Signs (INAS) [29]. The INAS assesses the presence vs. absence of the following neurological symptoms: hyperreflexia, areflexia, extensor plantar reflex, spasticity, paresis, muscle atrophy, fasciculations, myoclonus, rigidity, chorea/dyskinesia, dystonia, resting tremor, sensory symptoms, urinary dysfunction, cognitive dysfunction and brainstem oculomotor signs, the latter comprising ophthalmoparesis on horizontal and/or vertical gaze and/or slowing of saccades. The INAS count sums up the number of present additional neurological symptoms, ranging from 0 (no other neurological symptom) to a maximum of 16 neurological symptoms.
Age of ataxia onset was defined as the reported first occurrence of gait disturbances. For SCA3 mutation carriers, not yet experiencing gait disturbances or with missing information about the age of onset (n = 16), the age of onset was calculated on the basis of CAG repeat and age [30].
Mutation carriers with a SARA sum score of < 3 at baseline were categorized as pre-ataxic, according to the established SARA sum score cut-off, irrespective of SARA scores at follow-up visits. To assess the clinical phenotype, we examined the SCA3 mutation carrier within their haplotype at bp987 and bp1118.
Genetic analysis
The CAG repeat of ATXN3 was amplified using CAG repeat flanking primers [12]. Shortly, 150 ng of genomic DNA were amplified by a standard polymerase chain reaction (PCR) followed by capillary electrophoresis using the Beckman Coulter Fragment Analysis Software (Beckman Coulter).
SNPs at bp987 and bp1118 were determined in an allele-specific manner using fragment analyses. Therefore, PCR reactions were carried out using two different reverse primers that differ at the respective base position of the SNP bp 987 or bp1118, each labeled with a different fluorophore (forward primer 5′-CCAGTGACTACTTTGATTCG-3′ for both bp987 and bp1118 SNPs; reverse primer bp987 wildtype 5′-IRD700-ACTCTGTCCTGATAGGTCCCC−3′; reverse primer bp987 SNP 5′-Cy5-ACTCTGTCCTGATAGGTCCCG−3′; reverse primer bp1118 wildtype 5′-IRD700-GCAAAAATCACATGGAGCTCG−3′; reverse primer bp1118 SNP 5′-Cy5-GCAAAAATCACATGGAGCTCT−3′). 150 ng of genomic DNA was amplified in a standard PCR reaction containing 5% HiDi™ formamide (bp987; Life Technologies LTD, Warrington, UK) or 4 µl 5 × Q-Solution (bp1118; Qiagen, Hilden, Germany), respectively, and 10 µM of each primer. The optimal annealing temperatures were set at 59.5 °C for bp987 and 58 °C for bp1118, respectively. PCR products were separated by size using capillary electrophoresis determining the respective fluorophores using Beckman Coulter Fragment Analysis Software (Beckman Coulter, Brea, US).
Statistical analysis
Statistical analysis was performed with R (version 4.2.1), except for the pie-charts that were created with SPSS (Version 23.0.0.2). The annual SARA progression was calculated as the average annual rate between SARA at baseline and the last visit. Chi-square test was used to determine Hardy–Weinberg equilibrium (HWE).
We applied the exact Fisher test to study the distributions of SNPs at bp987 and bp1118 between the wildtype and the expanded alleles of SCA3 mutation carriers and healthy controls and also between ataxic and pre-ataxic SCA3 mutation carriers.
To study group differences between the subgroups of controls and SCA3 mutation carriers, and pre-ataxic and ataxic SCA3 mutation carriers, the exact Fisher test was used for the variable sex, and the Mann–Whitney test for the variables CAG repeat, SARA sum score, SARA-annual progression, INAS count and disease duration at baseline. Group differences of age at baseline and age of onset were determined using a T-test.
The relation between the four different haplotypes of the expanded allele within the group of ataxic SCA3 mutation carriers was tested with the Fisher’s test for sex, One-way ANOVA for age at baseline, reported and age of onset. Kruskal–Wallis Test was used for the variables CAG repeat length, SARA sum score, SARA-annual progression, INAS count and disease duration at baseline. Additionally, closed post-hoc test was done for further analysis of SARA sum score.
Furthermore, an exact Fisher test was used to compare the presence and absence of single INAS items between the four different haplotypes of the expanded allele within the group of ataxic SCA3 mutation carriers. Subsequently, those items that showed a significant difference, were subjected to a closed post-hoc test with a corrected alpha, due to alpha error correction for , with n equals the number of considered items, to study the relation of haplotypes and the respective INAS items.
Results
Cohort characterization
A total of 226 ataxic SCA3 mutation carriers, 54 pre-ataxic SCA3 mutation carriers and 113 healthy controls were included in the final analysis. Table 1 gives an overview of the demographic, genetic and clinical data of controls and all SCA3 mutation carriers. Eight subjects (4 SCA3 mutation carriers and 4 healthy controls) were excluded from the final analysis due to missing clinical data.
Table 1.
Clinical and demographic characteristics of SCA3 mutation carriers and healthy controls
| Controls | SCA3 | Sign | p value | |
|---|---|---|---|---|
| N | 113 | 280 | n.a | |
| N male/female (%) | 50/63 (44.2/55.8) | 138/142 (49.3/50.7) | 0.381 | |
|
Age at baseline Mean [SD] |
45.5 [14.2] | 48.2 [13.0] | 0.072 | |
|
CAG repeats longer allele Median [IQR] |
24.0 [23.0, 27.0] | 69.0 [66.0, 71.0] | ** | < 0.0013 |
|
Age of onset Mean [SD] |
n.a | 40.35 [9.78] | n.a | |
|
SARA sum score Median [IQR] |
0.0 [0.0, 0.50] | 9.5 [4.0, 17.0] | ** | < 0.0013 |
|
SARA sum score annual progression Median [IQR] |
0.0 [0.0, 0.0] | 0.81 [0.0, 2.1] | ** | < 0.0013 |
|
INAS count Median [IQR] |
1.0 [0.0, 1.0] | 4.5 [3.0, 7.0] | ** | < 0.0013 |
|
Disease duration at baseline, in years4 Median [IQR] |
n.a | 7.9 [3.6, 14.6] | n.a |
n.a. not applicable, sign. Significance, IQR interquartile range, SD standard deviation
*p < 0.05; **p < 0.001
1Fisher Test
2t-Test
3Mann-Whitney Test
4Disease duration is defined as the time from the age of onset (detailed description is given in the Methods section) until baseline visit, in years
The two patients who were homozygous for the disease-causing gene with two expanded alleles were excluded from the main analysis. Their demographic, genetic and clinical information is given separately.
Frequencies of both, the whole cohort (including healthy controls and mutations carriers) and control group, were in Hardy–Weinberg equilibrium (SNP 987: all p = 0.5998; CNTR p = 0.144; SNP 1118: all p = 0.158; CNTR p = 0.718) Deviation from Hardy–Weinberg equilibrium were found in mutation carriers for all analyzed SNPs (SNP 987 p = 0.0086; SNP 1118 p = 0.0144).
Distribution and frequency of SNPs at bp987 and bp1118 on the wildtype and expanded ATXN3 alleles
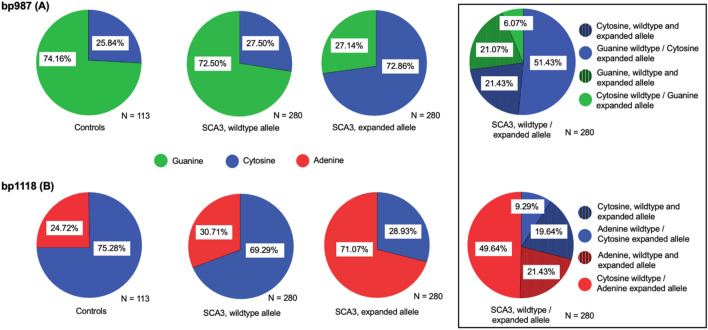
The distribution of the wildtype alleles of the SCA3 mutation carriers did not differ statistically from healthy controls. However, the distributions of SNPs in healthy controls and wildtype allele in SCA3 is significantly different from those observed on the expanded allele in SCA3 (p < 0.001, Fig. 1, Supplementary Table 1). For the SNP at position bp987 the majority of healthy controls and the majority of wildtype alleles in SCA3 showed Guanine (HC: 74.16%; wildtype allele SCA3: 72.5%), compared to only one-fourth of the expanded alleles in SCA3 (27.14%). For the SNP at bp1118 the majority of healthy controls as well as wildtype alleles in SCA3 showed Cytosine (HC: 75.28%, wildtype SCA3: 69.29%), compared to only 28.93% of the expanded alleles in SCA3 (Fig. 1). Genotype distribution in expanded alleles in ataxic and pre-ataxic SCA3 mutation carriers did not differ (Supplementary Table 1).
Fig. 1.
Relative frequencies of the SNP distributions at bp987 and bp1118 on the wildtype and expanded alleles. The relative frequencies of Guanine (green) and Cytosine (blue) at bp987 (A) and of Adenine (red) and Cytosine (blue) at bp1118 (B) are given for healthy controls as well as the wildtype and expanded allele in SCA3 mutation carriers, respectively. In the box on the far left, the distribution of the extended allele is further categorised: The respective proportion in which the wildtype and the expanded allele do not differ, e.g. do have the same nucleobase, is hatched. The distribution in controls is comparable to those of the wildtype allele in SCA3, while both are significantly different from the distribution on the expanded allele of SCA3 (p < 0.001)
Moreover, we studied the percentage, where the SCA3 wildtype allele differed from the expanded allele: In 57.5% the wildtype SNP differed from the expanded allele at bp 987, and in 58.9% at bp1118 (Fig. 1, right box).
The two homozygous SCA3 mutation carriers that were excluded from the final analysis had an early age of onset (23 and 30 years) and a high SARA sum score at baseline examination (28.5 and 23) with a disease duration of 16.05 and 17.02 years, respectively. They showed Cytosine—Guanine and Guanine—Guanine at bp987, and both Adenine—Adenine at bp1118. The CAG-repeats on the expanded alleles were 64/64 and 60/66, respectively.
Distribution of the SNPs at wildtype and expanded allele per research center
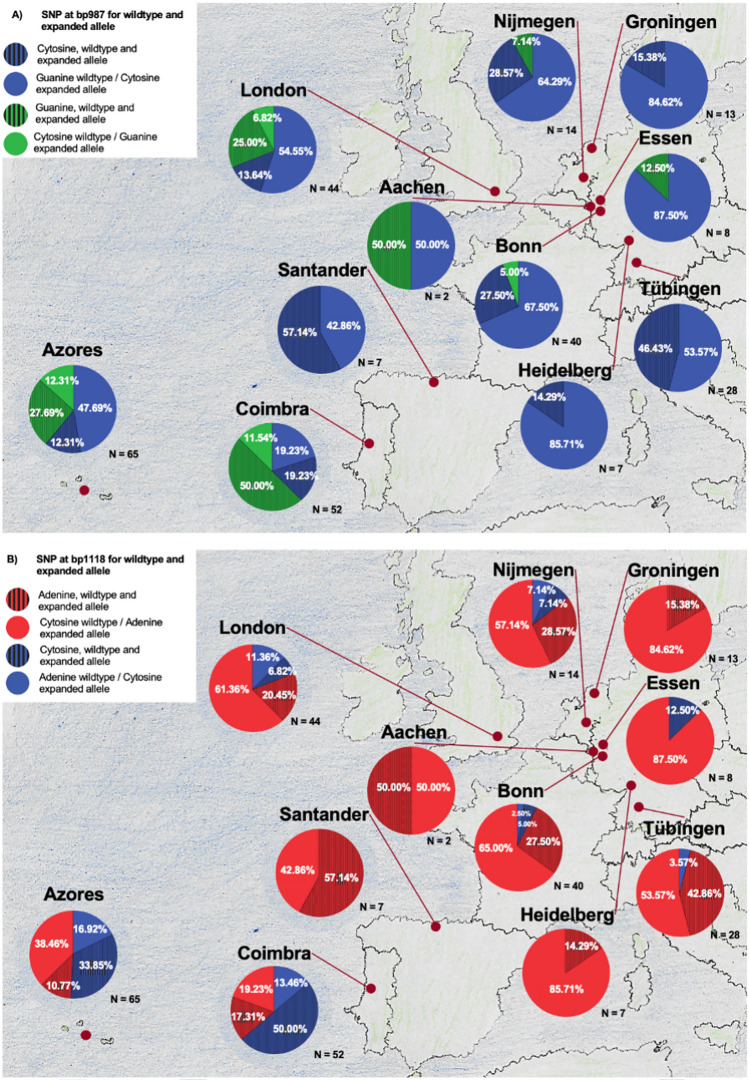
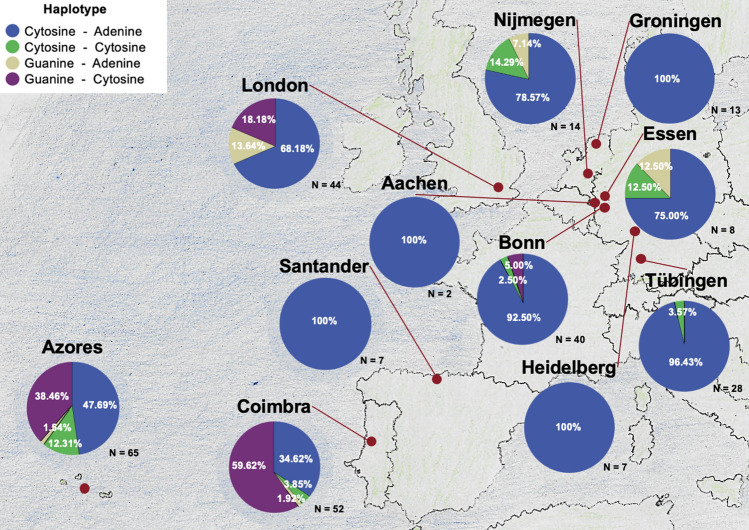
The majority of SCA3 mutation carriers at continental European research centers show on the expanded allele at bp987 Cytosine and Guanine on the wildtype. However, Portuguese research centers in Coimbra and on the Azores as well as London showed a different vice versa pattern. A similar constellation is depicted for the SNP at bp1118 with mostly Adenine on the expanded allele and Cytosine on the wildtype allele. Again, the Portuguese research centers in Coimbra and on the Azores showed a larger proportion of Cytosin on the expanded allele (Fig. 2). At the German site Aachen, only 2 participants were included, thus we do not recognize the proportions here as a trend.
Fig. 2.
Relative frequencies of the SNPs at bp987 (A) and bp1118 (B) of the wildtype and expanded allele for each European site. Color codes the nucleobase at the expanded allele: cytosine in blue, guanine in green and adenine in red. Full-colored areas indicate the proportion with a different nucleobase at wildtype and expanded allele, while the hatched areas indicate the presence of the same nucleobase at wildtype and expanded allele
Distribution of haplotypes and association with demographic, genetic and clinical data
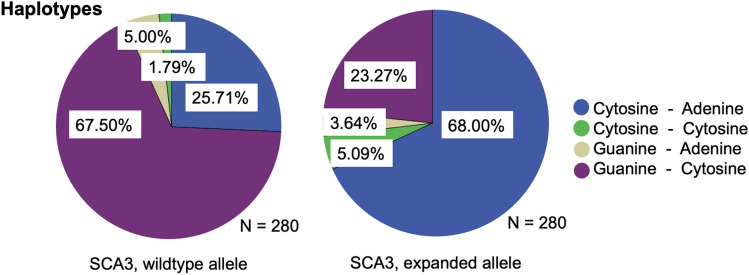
Four different haplotypes, defined by the SNPs at positions bp987 and bp1118, were found in both wildtype and expanded alleles of the analyzed SCA3 mutation carriers. The haplotypes of the wildtype allele in SCA3 mutation carriers present a frequency of 67.5% Guanine–Cytosine, 25.71% Cytosine–Adenine, 5.0% Guanine–Adenine and 1.79% Cytosine–Cytosine at bp987 and bp1118, while expanded alleles in SCA3 mutation carriers at bp987 and bp1118 demonstrate a frequency of 68.0% Cytosine–Adenine, 23.27% Guanine–Cytosine, 5.09% Cytosine–Cytosine and 3.64% Guanine–Adenine (Fig. 3).
Fig. 3.
Relative frequencies of the haplotypes of the wildtype and expanded allele at bp987 and bp1118 for SCA3 mutation carriers. Blue areas represent the frequency of Cytosine (bp987)–Adenine (bp1118), green areas the frequency of Cytosine–Cytosine, ochre areas the frequency of Guanine–Adenine and purple areas the frequency of Guanine–Cytosine, respectively
Genotypic and clinical evaluation of the cohort showed significant differences between SCA3 mutation carriers and controls in CAG repeat lengths, SARA sum score and annual progression and INAS count (p < 0.001, Table 1). Pre-ataxic and ataxic SCA3 mutation carriers showed significant differences regarding age at baseline, SARA sum score and SARA progression, INAS count as well as the disease duration at baseline (p < 0.001, supplementary Table 2).
SARA annual progression, INAS count, age of disease onset and disease duration at baseline as well as the CAG repeat length of the longer allele were similar between the four haplotypes in SCA3 (Table 2, supplementary Table 3).
Table 2.
Clinical and demographic characteristics of SCA3 mutation carriers for each haplotype of the expanded allele (bp987 and bp1118)
| Haplotype, expanded allele bp987–bp1118 | Sign | p value | ||||
|---|---|---|---|---|---|---|
| Cytosine–Adenine | Guanine–Cytosine | Cytosine–Cytosine | Guanine–Adenine | |||
| N | 189 | 66 | 15 | 10 | n.a | |
| N male/female (%) | 96/93 (50.8/49.2) | 30/36 (45.5/54.5) | 8/7 (53.3/46.7) | 4/6 (40/60) | 0.801 | |
|
Age at baseline Mean [SD] |
48.14 [12.41] | 48.23 [14.24] | 45.83 [12.72] | 51.87 [16.14] | 0.732 | |
|
CAG repeats longer allele Median [IQR] |
69.00 [66.00, 71.00] | 70.00 [66.00, 71.75] | 70.00 [67.50, 72.00] | 67.50 [66.25, 70.00] | 0.613 | |
|
Age of onset Mean [SD] |
40.75 [9.36] | 40.64 [10.84] | 36.67 [6.77] | 36.20 [13.16] | 0.232 | |
|
SARA sum score Median [IQR] |
9.50 [4.00, 16.00] | 9.00 [4.00, 17.25] | 13.50 [8.00, 23.75] | 20.00 [15.62, 22.25] | * | 0.013 |
|
SARA sum score annual progression Median [IQR] |
0.86 [0.00, 1.98] | 0.56 [− 0.38, 2.19] | 2.76 [0.52, 4.07] | 0.66 [0.28, 1.67] | 0.213 | |
|
INAS count Median [IQR] |
4.00 [2.00, 7.00] | 5.00 [3.00, 6.75] | 4.00 [3.50, 5.50] | 8.00 [5.50, 9.00] | 0.093 | |
|
Disease duration at baseline, in years4 Median [IQR] |
8.03 [3.40, 13.84] | 8.36 [2.98, 13.52] | 10.87 [5.37, 18.77] | 17.50 [12.05, 21.99] | 0.593 | |
n.a. non applicable, sign. Significance, IQR interquatile range, SD standard deviation; *p < 0.05
1Fisher-Test
2One-way Anova
3Kruskal–Wallis Test
4Disease duration is defined as the time from the age of onset (detailed description is given in the Methods section) until baseline visit, in years
SARA sum score in the group of all SCA3 mutation carriers (Table 2) and the subgroup of ataxic SCA3 mutation carriers (supplementary Table 3) showed a statistically significant difference between the haplotypes (p < 0.05; Table 2; supplementary Table 3). The post hoc testing revealed a statistically higher mean SARA sum score in ataxic SCA3 carriers with the haplotype Guanine—Adenine (mean SARA = 20.00, N = 10) and the haplotype Cytosine—Adenine (mean SARA 11.00, N = 150) (Supplementary Fig. 1 and Supplementary Table 3).
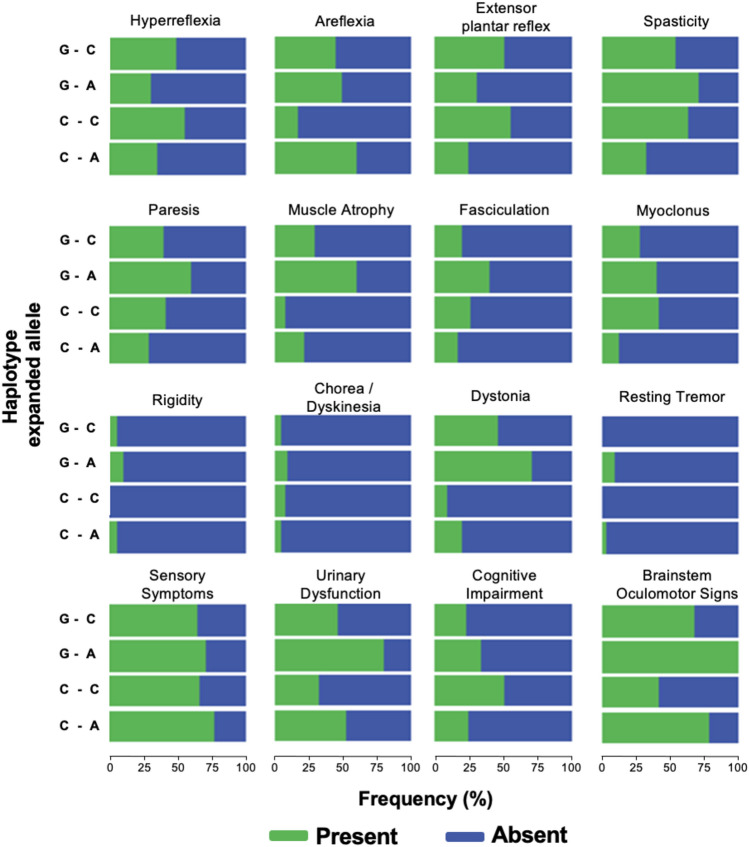
To further study potentially different phenotypes related to the analyzed SNPs we compared the distribution of INAS subitems among the haplotypes of the expanded allele in the group of ataxic SCA3 mutation carriers (Fig. 4; supplementary Table 4). Areflexia (C–A versus C–C), extensor plantar reflex (C–A versus G–C), spasticity (C–A versus G–C), muscle atrophy (C–A versus G–A), myoclonus, dystonia (C–A versus G–C, C–A versus G–A, C–C versus G–A) and brainstem oculomotor signs (C–A versus C–C; C–C versus G–A) showed a significant difference between the four haplotypes of the expanded allele (Supplementary Tables 4 and 5).
Fig. 4.
Relative frequencies of neurological, non-ataxia signs. The relative frequencies of additional neurological signs other than ataxia are given for the four haplotypes of the expanded allele (bp987 and bp1118) respectively: Guanine–Cytosine (G–C, N = 54), Guanine–Adenine (G–A, N = 10), Cytosine–Cytosine (C–C, N = 12) and Cytosine–Adenine (C–A, N = 150). Bars indicating the presence of each item are colored in green, and the remaining proportion with absence of the respective sign is marked in blue
Distribution of the haplotypes of the expanded allele per research center
To study the regional distribution in Europe, frequencies of haplotypes of the SCA3 expanded allele at bp987 and bp1118 were calculated for each research center separately (Fig. 5). In Portugal, SCA3 mutation carriers from the Azores (N = 65) and from Coimbra, mainland Portugal (N = 52) showed all possible haplotypes with a particularly high proportion of G–C (Fig. 5, Supplementary Table 6).
Fig. 5.
Relative frequencies of haplotypes at SNP bp987–bp1118 on the expanded allele for each European site. The blue area represents the frequency of Cytosine (bp987) –Adenine (bp1118), the green area the frequency of Cytosine–Cytosine, the ochre area the frequency of Guanine–Adenine and the purple area the frequency of Guanine–Cytosine, respectively
Discussion
To explore the potential of well-known SNPs (bp987, bp1118) in ATXN3 for allele-specific therapies, such as ASOs, we studied the frequency and regional distribution in a large European cohort of > 200 SCA3 mutation carriers. In line with previous studies, the distribution of haplotypes on the expanded allele showed a considerable difference from those of wildtype alleles of SCA3 mutation carriers and of healthy controls. Additionally, this distribution across Europe was mainly influenced by regional origin. The haplotype of the two intragenic SNPs at bp987 and bp1118 is known to be linked specifically to the CAG expansion in around 70–72% of the mutation carriers [4, 21, 22]. Similar results were found in our study in which 68% of the expanded alleles presented the haplotype C-A at bp987-1118, whereas 67.5% of the wildtype alleles in mutation carriers showed the G-C haplotype. Our comprehensive European haplotyping is in line with previous smaller or less regional distributed descriptions of haplotypes in SCA3 [21]. While the eight German, Dutch and Spanish research centres exhibited limited to no variation with a predominance of the C–A haplotype, associated to the Joseph lineage, of 92.8% to 100%, this haplotype was less frequent in the UK (68%), the Azores (48%) and the Portugal mainland (35%). In contrast, the latter three research centres showed an increased proportion of the Haplotype G–C (18.2%, 38.5% and 59.6%, respectively), associated to the Machado lineage. The haplotypes C–C and G–A were found less frequent with a specific regional origin. In previous studies, the G–A haplotype was described in one family from Morocco, French Guyana and two families from the United States. In addition, the C–C haplotype was not described earlier in SCA3 mutation carriers [21].
Our results provide for the first time a comprehensive overview of the European distribution, where regional differences may partially be explained by different main founder mutations. As highly standardized clinical data were available, we could for the first time analyse the relation between haplotypes and the clinical phenotype. Our data demonstrated, that the SARA sum score and the distribution of INAS subitems were slightly significantly differently distributed among the different haplotypes. Generally, patients with the most common haplotype C–A at the expanded allele demonstrated less frequently pathological extensor plantar reflexes and spasticity compared to the second most often haplotype G–C. Importantly, SCA3 mutation carriers carrying haplotypes C–A and G–C had similar numbers of CAG repeats in the expanded allele, providing evidence that this effect is not biased by different repeat length. Even though, differences in haplotype expression are conceivably related to different phenotypes and symptoms due to ATXN3 isoforms caused by SNPs. It is well known that the SNP at bp1118 leads to a premature stop codon and therefore, to a shorter ATXN3 isoform, known as ATXN3a short (ATXN3aS) [31]. The two most expressed ATXN3 isoforms in the brain ATXN3a short (ATXN3aS, harbouring the A at bp1118) and ATXN3a long (ATXN3aL, harbouring the wildtype sequence with C at bp1118) [32], demonstrated that the isoform ATXN3aS showed shorter half-life, faster degradation by the autophagic as well as proteasomal pathways and formation of larger aggregates compared to ATXN3aL [31]. However, in summary, we could not identify reliable overarching clusters of phenotypic features related to one haplotype. Nevertheless, the haplotypes C–C and G–A were underrepresented, thus further studies with an increased number of SCA3 mutation carriers, in particular of those haplotypes, are needed.
Surprisingly, genotype analyses revealed that only around 60% of SCA3 mutation carriers will benefit from an allele-specific protein-lowering therapy as 40% harbours the same nucleobase at the wildtype and the expanded allele. ASOs and siRNAs aiming at silencing the ATXN3 gene are currently promising therapeutic options in SCA3 [12–17]. Because of autosomal-dominantly inheritance, most SCA3 mutation carrier represent only one pathogenic expanded allele, therefore an allele-specific gene therapy approach is of particular interest to preserve the physiological function of the wildtype allele [31, 33]. Allele-specificity can be achieved by using SNPs that are specifically linked to the expanded allele. As ASOs and siRNA are short (12–25 nucleotides long) in size, only one or two very closely located SNPs can be targeted by this allele-specific strategy [32, 33, 34, 35]. Therefore, a haplotype-based stratification strategy is not helpful to develop new allele-specific therapies. To get more insights into the use of SNP for allele-specific targeting, we especially focused on the genotype of SNP 987 and 1118 in our large European SCA3 cohort. On average our data with approximately 68% are in line with previous reports of 70–72% of Cytosine for bp987 and Adenine for bp1118 [4, 21, 22, 25]. However, the proportion of SCA3 mutation carriers which are non-treatable with an allele-specific therapy considering the SNPs bp987 and bp1118 since their nucleobases do not differ between wild type and expanded allele differs strongly depending mainly on the origin (in our study between 12 and 69%). There will be SCA3 mutation carrier (in our study between 12 and 69%, depending on the regional origin) which are non-treatable with an allele-specific therapy considering SNPs at bp987 and bp1118 since their nucleobases do not differ between wildtype and expanded allele. Therefore, there is an urgent need to further characterize the allele-specificity of other ATXN3 specific SNPs in large, regional wide-spread SCA3 cohorts. In summary, with allele-specific protein-lowering therapies under investigation, there is an urgent need to understand the worldwide distribution of SNPs linked to the expanded allele in SCA3. Currently, SNP determination within ATXN3 is not part of the standard diagnostic genetic CAG repeat evaluation. Our study provided important information about the European distribution of the two examined SNPs, bp987 and bp1118, and might therefore be of help by providing a trial-ready cohort for allele-specific, protein-lowering therapies.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The ESMI consortium would like to thank Ruth Herberz and Nina Roy-Kluth for the coordination and management of the ESMI project.
ESMI study group members: Dagmar Timmann, JARA-BRAIN Institute Molecular Neuroscience and Neuroimaging, Research Centre Juelich GmbH and RWTH Aachen University, Aachen, Germany; Andreas Thieme, JARA-BRAIN Institute Molecular Neuroscience and Neuroimaging, Research Centre Juelich GmbH and RWTH Aachen University, Aachen, Germany.
Abbreviations
- SNPs
Single nucleotide polymorphism
- SCA3
Spinocerebellar ataxia type 3
- polyQ Tract
Polyglutamine tract
- ASO
Antisense oligonucleotide
- ANOVA
Analysis of variance
- C
Cytosine
- A
Adenine
- G
Guanine
- SARA
Scale for the assessment and rating of ataxia
- INAS
Inventory of non-ataxia signs
Author contributions
T.E.: Research Project: Organization; Statistical Analysis: Design, Execution; Manuscript Preparation: Writing of the first draft. D.S.: Research Project: Conception, Organization, Execution. M.M.S.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. T.S.: Statistical Analysis: Design, Execution, Review and Critique; Manuscript Preparation: Review and Critique. M.R.: Research Project: Conception; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. A.R.V.M.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. M.L.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. B.K.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. D.O.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. M.G.E.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. S.L.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. P.S.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. L.P.d.A.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. P.G.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. H.G.M.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. S.N.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. J.d.V.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. B.P.v.d.W.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. J.v.G.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. M.S.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. L.S.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. K.R.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. F.E.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. H.J.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. J.I.: Research Project: Organization; Statistical Analysis: Review and Critique; Manuscript Preparation: Review and Critique. O.R. Research Project: Conception, Organization; Manuscript Preparation: Review and Critique. T.K.: Statistical Analysis: Design, Review and Critique; Manuscript Preparation: Review and Critique. J.F.: Statistical Analysis: Design, Review and Critique; Manuscript Preparation: Writing of the first draft. J.H.S.: Research Project: Conception, Organization; Manuscript Preparation: Writing of the first draft.
Funding
Open Access funding enabled and organized by Projekt DEAL. This publication is an outcome of ESMI, an EU Joint Programme—Neurodegenerative Disease Research (JPND) project (see www.jpnd.eu). The project is supported through the following funding organisations under the aegis of JPND: Germany, Federal Ministry of Education and Research (BMBF; funding codes 01ED1602A/B); Netherlands, The Netherlands Organisation for Health Research and Development; Portugal, Fundação para a Ciência e a Tecnologia (FCT); United Kingdom, Medical Research Council.
Availability of data and materials
Data will be made available upon reasonable request and as patient consent allow.
Declarations
Conflicts of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
The study was approved by the local ethics committees of all participating centers. Informed and written consent was obtained from all study participants at enrollment.
Consent for publication
Not applicable.
Footnotes
Tim Lukas Elter, Daniel Sturm, Jennifer Faber and Jeannette Hübener-Schmid have contributed equally to this work.
Contributor Information
Tim Lukas Elter, Email: Tim-lukas.elter@dzne.de, Email: timlukaselter@gmail.com.
ESMI study group:
References
- 1.Klockgether T, Mariotti C, Paulson HL (2019) Spinocerebellar ataxia. Nat Rev Dis Primers 5(1):24 [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt C, Lima M (2011) Machado-Joseph Disease: from first descriptions to new perspectives. Orphanet J Rare Dis 6:35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo M, Raposo M, Kazachkova N, Vasconcelos J, Kay T, Lima M (2016) Trends in the epidemiology of spinocerebellar ataxia type 3/machado-joseph disease in the azores islands. Portugal JSM Brain Science 1:1–5 [Google Scholar]
- 4.Maciel P, Gaspar C, Guimarães L, Goto J, Lopes-Cendes I, Hayes S et al (1999) Study of three intragenic polymorphisms in the Machado-Joseph disease gene (MJD1) in relation to genetic instability of the (CAG)n tract. Eur J Hum Genet 7(2):147–156 [DOI] [PubMed] [Google Scholar]
- 5.Ichikawa Y, Goto J, Hattori M, Toyoda A, Ishii K, Jeong SY et al (2001) The genomic structure and expression of MJD, the Machado-Joseph disease gene. J Hum Genet 46(7):413–422 [DOI] [PubMed] [Google Scholar]
- 6.Takiyama Y, Nishizawa M, Tanaka H, Kawashima S, Sakamoto H, Karube Y et al (1993) The gene for Machado-Joseph disease maps to human chromosome 14q. Nat Genet 4(3):300–304 [DOI] [PubMed] [Google Scholar]
- 7.Ashizawa T, Öz G, Paulson HL (2018) Spinocerebellar ataxias: prospects and challenges for therapy development. Nat Rev Neurol 14(10):590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sequeiros J, Seneca S, Martindale J (2010) Consensus and controversies in best practices for molecular genetic testing of spinocerebellar ataxias. Eur J Hum Genet 18(11):1188–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLoughlin HS, Moore LR, Paulson HL (2020) Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol Dis 134:104635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wullner U, Evert B, Klockgether T (1998) Mechanisms of cell death in cerebellar disorders. Restor Neurol Neurosci 13(1–2):69–73 [PubMed] [Google Scholar]
- 11.Da Silva JD, Teixeira-Castro A, Maciel P (2019) From pathogenesis to novel therapeutics for spinocerebellar ataxia type 3: evading potholes on the way to translation. Neurotherapeutics 16(4):1009–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hauser S, Helm J, Kraft M, Korneck M, Hübener-Schmid J, Schöls L (2022) Allele-specific targeting of mutant ataxin-3 by antisense oligonucleotides in SCA3-iPSC-derived neurons. Mol Ther Nucleic Acids 27:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLoughlin HS, Gundry K, Rainwater O, Schuster KH, Wellik IG, Zalon AJ et al (2023) Antisense oligonucleotide silencing reverses abnormal neurochemistry in spinocerebellar ataxia 3 mice. Ann Neurol 94(4):658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLoughlin HS, Moore LR, Chopra R, Komlo R, McKenzie M, Blumenstein KG et al (2018) Oligonucleotide therapy mitigates disease in spinocerebellar ataxia type 3 mice. Ann Neurol 84(1):64–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martier R, Sogorb-Gonzalez M, Stricker-Shaver J, Hübener-Schmid J, Keskin S, Klima J et al (2019) Development of an AAV-based microRNA gene therapy to treat Machado-Joseph disease. Mol Ther Methods Clin Dev 15:343–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL et al (2003) Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci USA 100(12):7195–7200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nóbrega C, Nascimento-Ferreira I, Onofre I, Albuquerque D, Hirai H, Déglon N et al (2013) Silencing mutant ataxin-3 rescues motor deficits and neuropathology in Machado-Joseph disease transgenic mice. PLoS One 8(1):e52396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Z, Chen Z, Wang C, Huang F, Peng H, Hou X et al (2015) Two novel SNPs in ATXN3 3′ UTR may decrease age at onset of SCA3/MJD in Chinese patients. PLoS One 10(2):e0117488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang XZ, Huang XY, Yao JG, Wang C, Xia Q, Long XD (2018) Genetic polymorphisms in ataxin-3 and liver cirrhosis risk related to aflatoxin B1. Oncotarget 9(44):27321–27332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh B, Bose P, Shamal SN, Joshi D, Singh R (2020) Polymorphic study of ataxin 3 gene in eastern Uttar Pradesh population. J Anatom Soc India 69(4):226–232 [Google Scholar]
- 21.Gaspar C, Lopes-Cendes I, Hayes S, Goto J, Arvidsson K, Dias A et al (2001) Ancestral origins of the Machado-Joseph disease mutation: a worldwide haplotype study. Am J Hum Genet 68(2):523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan SR, Ni W, Dong Y, Wang N, Wu ZY (2015) Population genetics and new insight into range of CAG repeats of spinocerebellar ataxia type 3 in the Han Chinese population. PLoS One 10(8):e0134405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopes SM, Faro R, Lopes MM, Onofre I, Mendonça N, Ribeiro J et al (2020) Protocol for the characterization of the Cytosine-Adenine-Guanine tract and flanking polymorphisms in Machado-Joseph Disease: impact on diagnosis and development of gene-based therapies. J Mol Diagn 22(6):782–793 [DOI] [PubMed] [Google Scholar]
- 24.Martins S, Calafell F, Gaspar C, Wong VC, Silveira I, Nicholson GA et al (2007) Asian origin for the worldwide-spread mutational event in Machado-Joseph disease. Arch Neurol 64(10):1502–1508 [DOI] [PubMed] [Google Scholar]
- 25.Martins S, Soong BW, Wong VC, Giunti P, Stevanin G, Ranum LP et al (2012) Mutational origin of Machado-Joseph disease in the Australian Aboriginal communities of Groote Eylandt and Yirrkala. Arch Neurol 69(6):746–751 [DOI] [PubMed] [Google Scholar]
- 26.Prudencio M, Garcia-Moreno H, Jansen-West KR, Al-Shaikh RH, Gendron TF, Heckman MG et al (2020) Toward allele-specific targeting therapy and pharmacodynamic marker for spinocerebellar ataxia type 3. Sci Transl Med. 10.1126/scitranslmed.abb7086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santana MM, Gaspar LS, Pinto MM, Silva P, Adao D, Pereira D et al (2023) A standardised protocol for blood and cerebrospinal fluid collection and processing for biomarker research in ataxia. Neuropathol Appl Neurobiol 49(2):e12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmitz-Hubsch T, du Montcel ST, Baliko L, Berciano J, Boesch S, Depondt C et al (2006) Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology 66(11):1717–1720 [DOI] [PubMed] [Google Scholar]
- 29.Jacobi H, Rakowicz M, Rola R, Fancellu R, Mariotti C, Charles P et al (2013) Inventory of Non-Ataxia Signs (INAS): validation of a new clinical assessment instrument. Cerebellum 12(3):418–428 [DOI] [PubMed] [Google Scholar]
- 30.Tezenas du Montcel S, Durr A, Rakowicz M, Nanetti L, Charles P, Sulek A et al (2014) Prediction of the age at onset in spinocerebellar ataxia type 1, 2, 3 and 6. J Med Genet 51(7):479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weishäupl D, Schneider J, Peixoto Pinheiro B, Ruess C, Dold SM, von Zweydorf F, et al (2019) Physiological and pathophysiological characteristics of ataxin-3 isoforms. J Biol Chem 294(2):644–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raposo M, Hübener-Schmid J, Tagett R, Ferreira AF, Melo ARV, Vasconcelos J, et al. Blood and cerebellar abundance of ATXN3 splice variants in spinocerebellar ataxia type 3/Machado-Joseph disease. bioRxiv. 2023:2023.04.22.537936 [DOI] [PubMed]
- 33.Matos CA, de Macedo-Ribeiro S, Carvalho AL (2011) Polyglutamine diseases: the special case of ataxin-3 and Machado-Joseph disease. Prog Neurobiol 95(1):26–48 [DOI] [PubMed] [Google Scholar]
- 34.Crooke ST, Baker BF, Crooke RM, Liang X-H (2021) Antisense technology: an overview and prospectus. Nat Rev Drug Discov 20(6):427–453 [DOI] [PubMed] [Google Scholar]
- 35.Siomi H, Siomi MC (2009) On the road to reading the RNA-interference code. Nature 457(7228):396–404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available upon reasonable request and as patient consent allow.