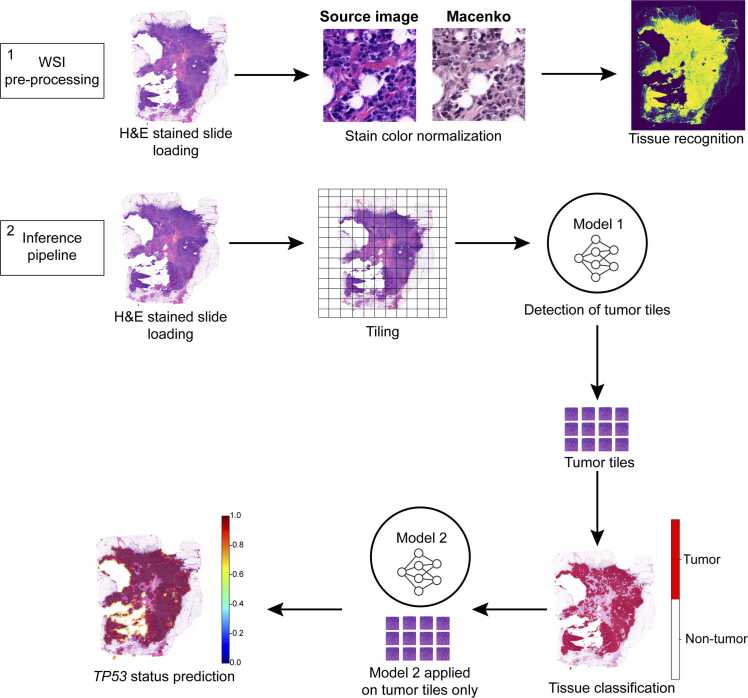
Fig. 1.
: Deep-learning pipeline workflow. The workflow begins with the pre-processing of whole slide images (WSI), which are first loaded into the Python environment. Once loaded, the images undergo stain-color normalization to standardize variations in staining, followed by the identification of relevant tissue areas for analysis. The inference pipeline then processes these images by segmenting them into smaller tiles. These tiles serve as the input for the first model, which is responsible for distinguishing between tumor and non-tumor regions. The classification results are visualized using a red-and-white mask, where red represents tumor areas and white represents non-tumor tissue. Finally, tiles identified as tumorous are further analyzed by a second model that predicts molecular features, such as TP53 mutation status, based on the whole image.