Figure 2.
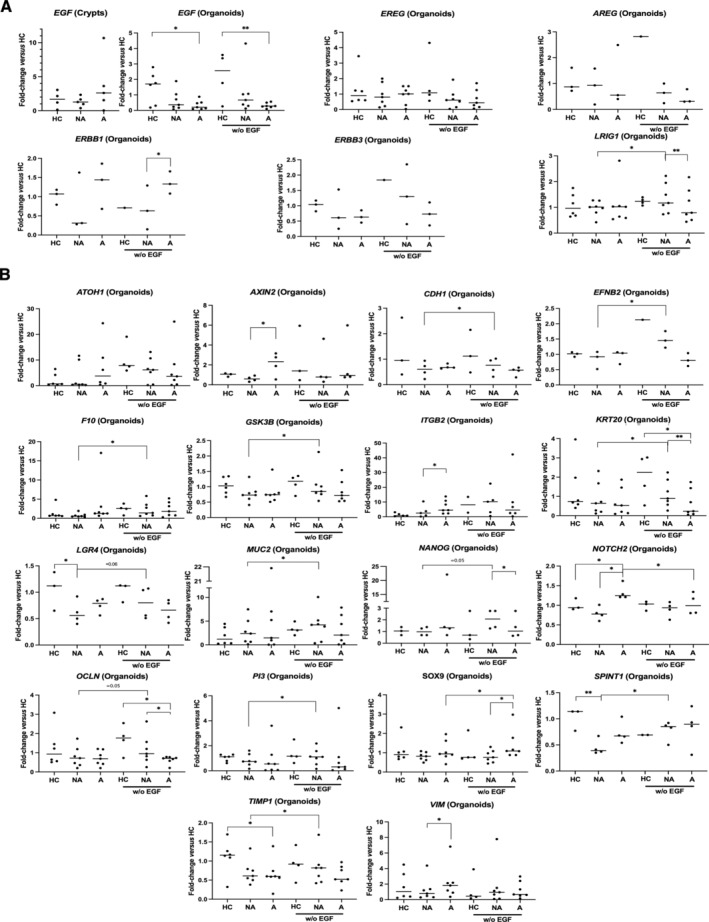
Transcriptional regulation by EGF in FAP organoids. (A) At D7 of culture, data on the EGFR pathway show a decrease of EGF mRNA in FAP organoids (NA, nonadenomatous; A, adenomatous) but not crypts compared to healthy controls (HCs). Under EGF starvation in the culture medium from D2, no change in the expression of several genes (EGF, EREG, AREG, ERBB1, ERBB3) was measured, except an increase for the EGFR regulator LRIG1 (NA organoids). (B) Starvation of EGF induced a loss of changes in the mRNA expression associated with the FAP phenotype: AXIN2, NOTCH2, VIM mRNA in A compared to NA and/or HC organoids; ITGB2, LGR4, SPINT1, TIMP1 mRNA in NA compared to A and/or HC organoids. In FAP organoids, EGF‐deprivation increased the expression of genes involved in crypt differentiation (SOX9, A organoids; ATOH1, CDH1, EFNB2, F10, GSK3B, KRT20, MUC2, NANOG, OCLN, PI3, NA organoids). n patients: crypts, HC = 4, NA/A = 5; organoids, HC = 3–6, NA/A = 3–7. ANOVA comparison between HC and NA/A: *p < 0.05 **p < 0.01, paired t‐test comparison between NA and A, NA and NA without EGF, A and A without EGF: *p < 0.05 **p < 0.01.
