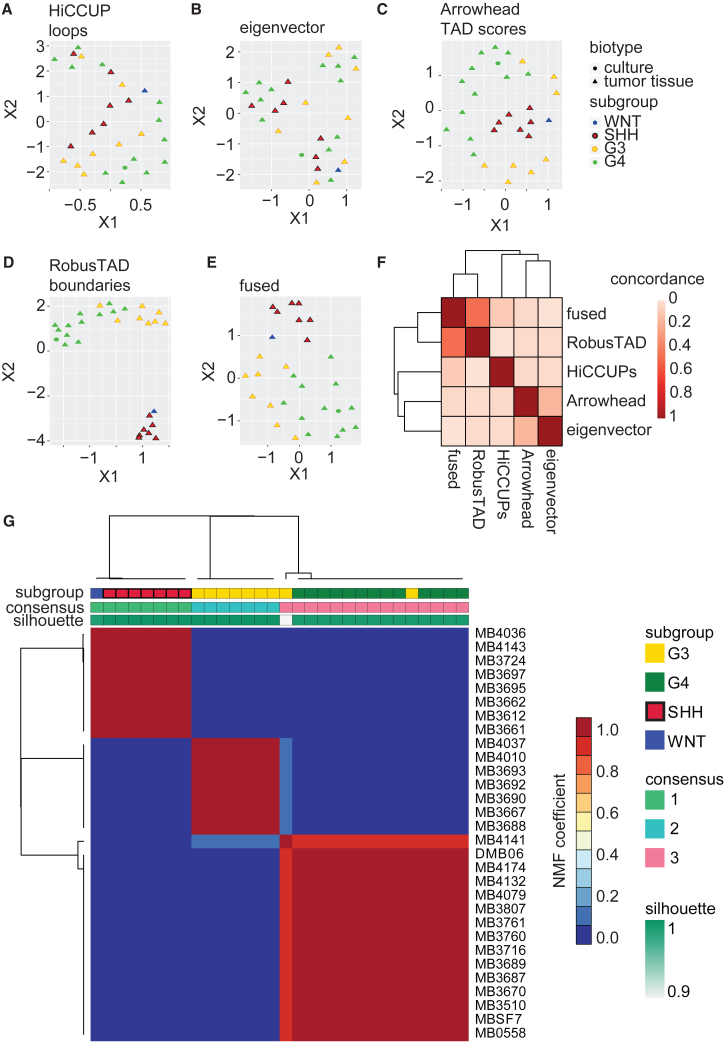
Figure 3.
TAD insulation scores distinguish MB molecular subgroups
(A) UMAP plot of MB samples based on loops called with HiCCUP.
(B) UMAP plot generated with eigenvector values, which provide information on compartments.
(C) UMAP plot generated with TAD scores called with Arrowhead.
(D) UMAP plot generated with boundary scores calculated with RobusTAD.
(E) UMAP plot based on fused distance metrics.
(F) Concordance between metrics derived from Hi-C data described in this study.
(G) Non-negative matrix factorization (NMF) analysis of TAD insulation scores for all MB samples, with k = 3. MB subgroups (G3, G4, SHH, and WNT) are indicated at the top of the image, highlighting the ability of TAD insulation scores to discriminate between subgroups. Consensus clusters are shown in green, blue, and pink. Silhouette scores provide the confidence level for the clustering calls for each sample.