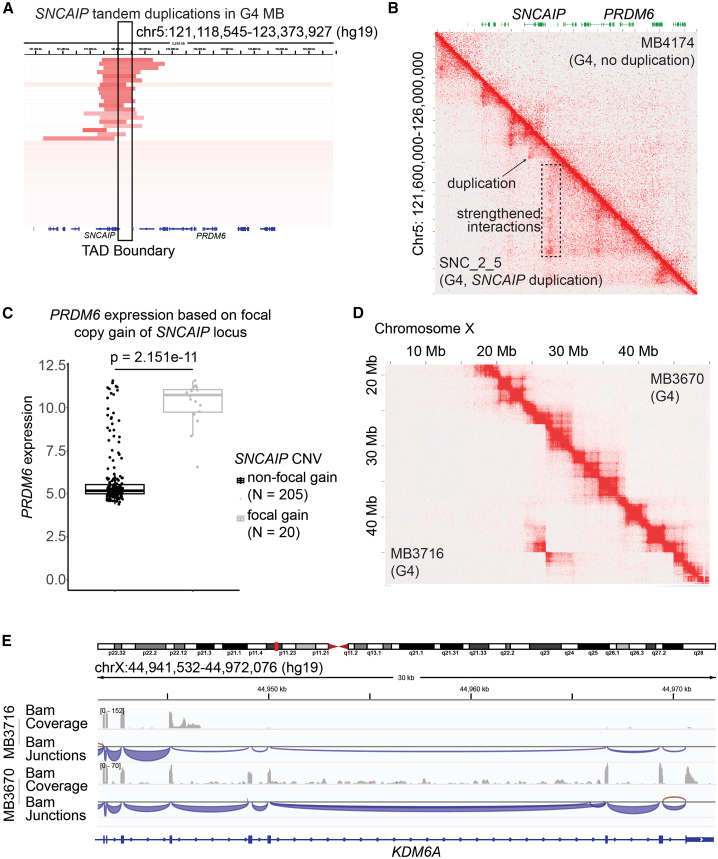
Figure 6.
Relationships between SVs, 3D genome, and transcriptional profiles
(A) Location of recurrent SNCAIP tandem duplications in the context of TAD boundaries in G4 MB. The coordinates of the region of chromosome 5 shown are indicated at the top. Copy number data for each MB sample are displayed along rows. MB samples with duplications are visible at the top of the diagram, with duplicated regions shown in red. The TAD boundary between the SNCAIP and PRDM6 loci is highlighted with a black rectangle.
(B) Observed minus expected (Obs-Exp) contact frequencies were determined by Hi-C data for a G4 MB with SNCAIP duplication (lower half) and for a G4 sample without the duplication (upper half). The genomic region represented is on chromosome 5 (coordinates shown along the y axis of the heatmap).
(C) PRDM6 transcriptional levels in samples with and without the PRDM6 duplication. p value was calculated with the two-tailed Mann Whitney U test.
(D) Inference of an SV affecting the KDM6A locus in a G4 MB (lower half of the Hi-C map). A G4 sample without SVs at this locus is displayed in the upper half of the heatmap.
(E) RNA-seq data were used to determine the effects of the SV at the KDM6A locus on the transcriptional levels of this gene.