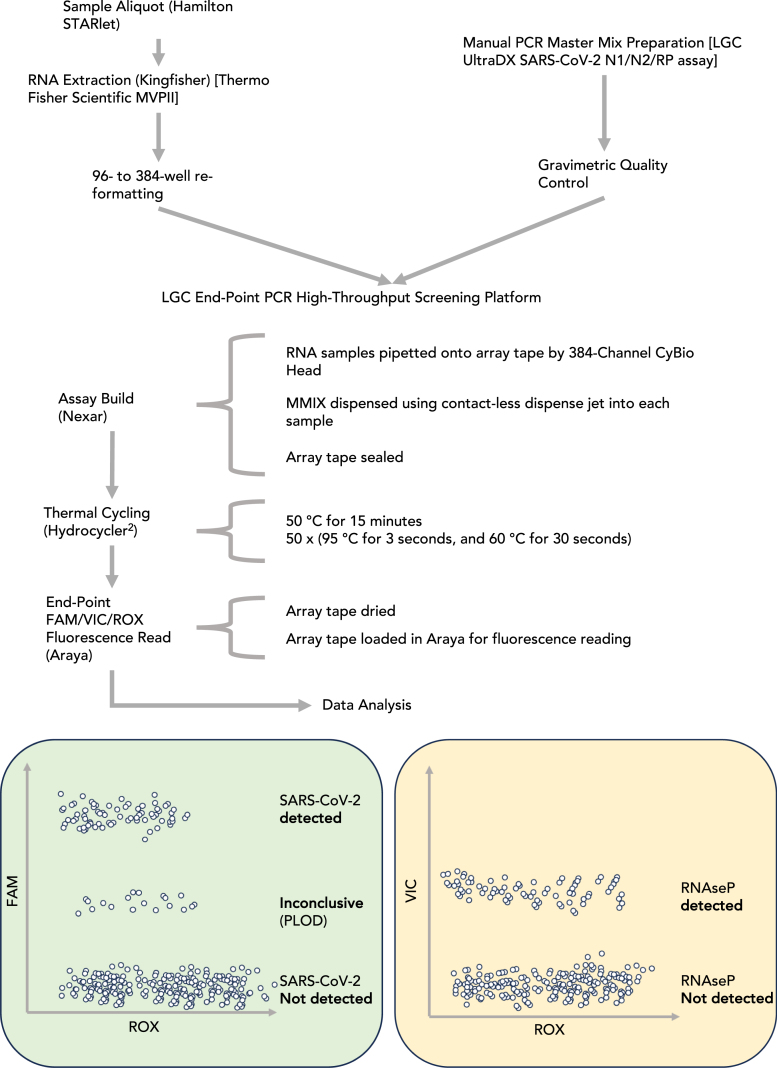
Fig. 1.
Process flow schematic for SARS-CoV-2 endpoint PCR (ePCR) workflow. Samples were purified using the Thermo Fisher Scientific MagMAX Viral Pathogen II kit and re-formatted from 96-deep-well to 384-well ANSI/SLAS microplates to prepare for assay build-in the Nexar Liquid handler 384-well Array Tape (AT). In parallel, PCR Master Mix (MMIX) was prepared and underwent a gravimetric quality control step before release for a run on the Nexar liquid handler. In practice, a run on the LGC ePCR High-throughput screening platform consisted of several (up to 16) 384-well microplates and a batch of PCR MMIX sufficient for all PCR reactions. The Nexar liquid handler first transfers samples with a 384-tip Dispense Pipet (DP) from the 384-well microplates to the AT before adding MMIX to each well of the tape with a Dispense Jet (DJ). The AT was then sealed, and reverse transcription and thermal cycling were conducted in a Hydrocycler2 (HC2) automated water bath. After the PCR reaction was completed, the Araya read the fluorescence of each well of the AT. Collected FAM (SARS-CoV-2 N1 and N2 targets) and VIC (human RNase P target) fluorescence data were normalized to ROX loading control and formed distinct clusters when plotted. The upper cluster in the FAM/ROX plot (green) was scored as SARS-CoV-2 detected, and occurs when both N1 and N2 targets amplify. The middle cluster, scored as Inconclusive or “Positive at Limit of Detection (PLOD),” refer to main text for further detail. The lower cluster was scored as SARS-CoV-2 Not Detected, corresponding with neither N1 nor N2 amplifying. The VIC/ROX plot (yellow) clusters into only two groups, scored as RNase P Detected, or RNase P Not Detected. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)