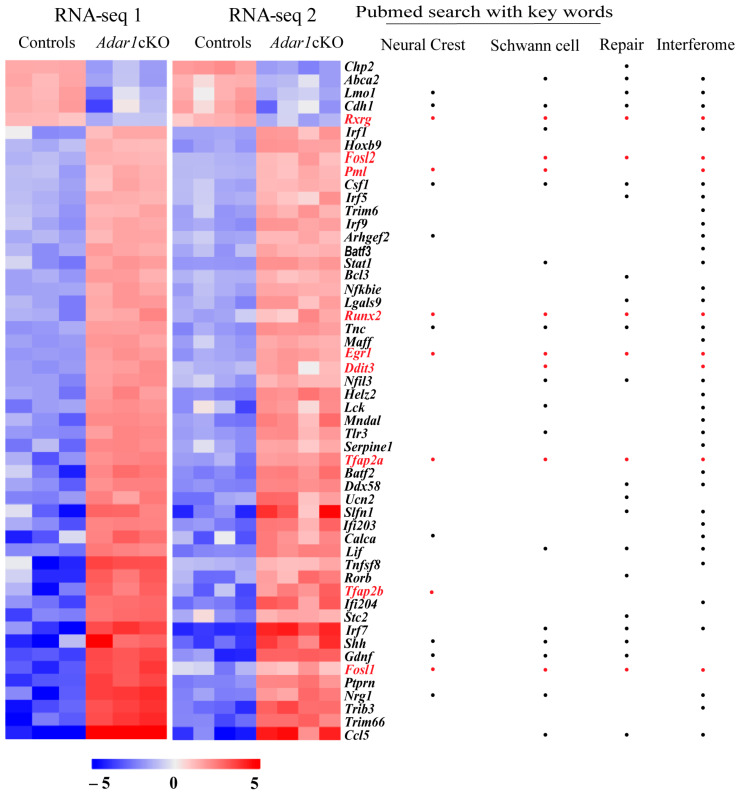
Figure 1.
Transcriptional regulator transcripts deregulated in the sciatic nerves of Adar1cKO versus the controls. Transcriptomic analysis identified 52 transcriptional regulators deregulated in the sciatic nerves of HtPA-Cre; Adar1fl/fl (Adar1cKO) mutants relative to the controls with fold change (FC ≥ 5). Heatmap representations of differentially-expressed transcriptional regulators [found in association with the GO terms mentioned in the text and deregulated more than fivefold in Adar1cKO mutants (n = 7) relative to the controls (n = 7) in RNA-Seq 1 and RNA-Seq 2 datasets]. The name of each gene is indicated on the right, with whether it belongs to the Interferome database, or involved in the Schwann cell repair process (see Table S3 for PMIDs). Genes found in the literature using the “neural crest” or “Schwann cells” keywords are also indicated. Colors reflect the z-score. Deregulated transcripts indicated in red (including dots) were the ones selected for further experimental validation.