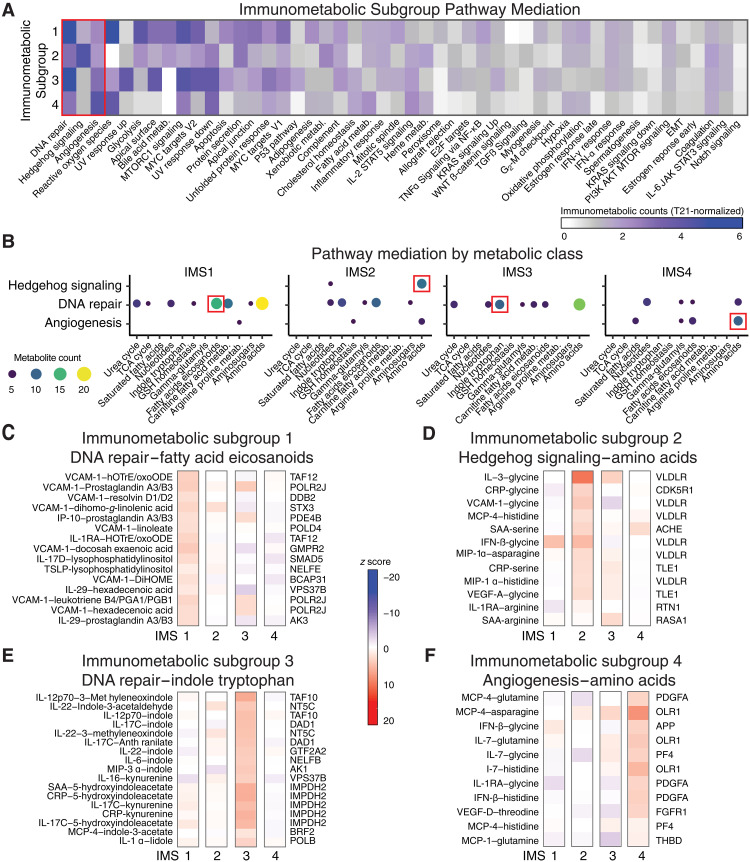
Fig. 5. Immunometabolic subgroups reveal differential mediation of cytokine-metabolite relationships.
(A) Number of immunometabolic relationships that were significantly (FDR < 0.05) and positively enriched (NES > 0) for mediation by genes in gene sets across each immunometabolic subgroup (IMS). Counts of enriched gene sets within each IMS were normalized to the counts of enriched gene sets across all individuals with T21. (B) Enrichment of metabolite classes for the cytokine-metabolite relationships mediated by the gene sets that were most differential across IMSs [red box in (A): DNA repair, Hedgehog signaling, and angiogenesis). (C to F) Top gene mediators and their relative strength of mediation (z scores) for cytokine-metabolite relationships within a selected metabolite class for each IMS [red boxes in (B)] are reported. (C) For IMS1, the top genes within DNA repair that mediate fatty acid/eicosaniod cytokine-metabolite relationships are shown. (D) For IMS2, the top genes within Hedgehog signaling that mediate amino acid cytokine-metabolite relationships are shown. (E) For IMS3, the top genes within DNA repair that mediate indole and tryptophan cytokine-metabolite relationships are shown. (F) For IMS4, the top genes within Angiogenesis that mediate amino acid cytokine-metabolite relationships are shown.