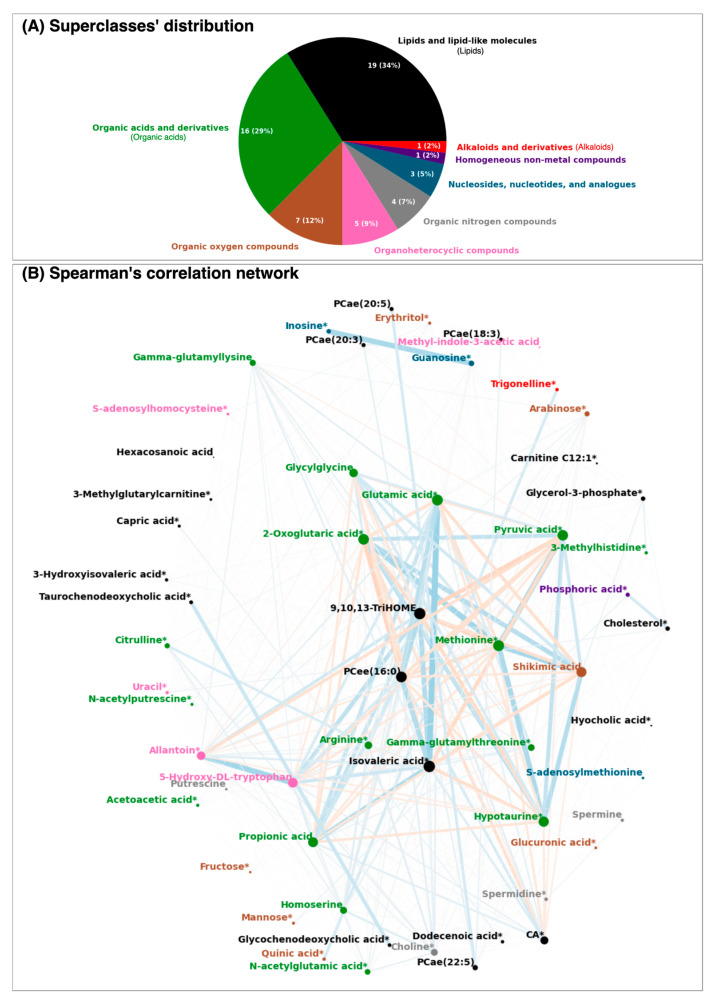
Figure 1.
The selected metabolites’ (A) superclass distribution and (B) Spearman’s correlation network. In the network, the nodes correspond to the metabolites and the edges depend on the strength of their Spearman’s correlation between two nodes. Thicker and darker edges indicate a higher pairwise correlation, whereas thinner and lighter colors indicate a lower correlation. Red edges correspond to negative correlations and blue edges to positive ones. The node colors specify the metabolite superclass, and its size increases according to the absolute strength of the edges connected to it. Metabolites marked with * represent those available also in the external validation dataset. For visualization purposes, the correlations were powered to 4 but kept the original signal. In this figure, CA stands for caproic acid.