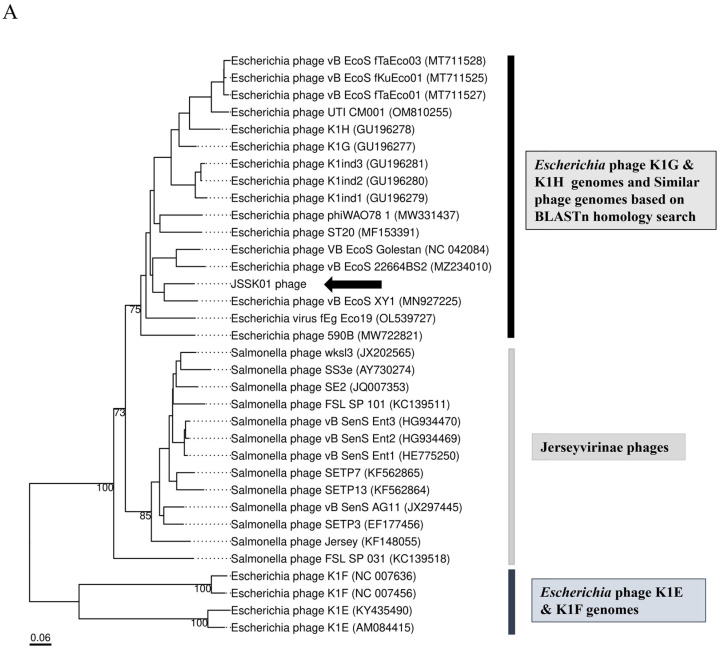
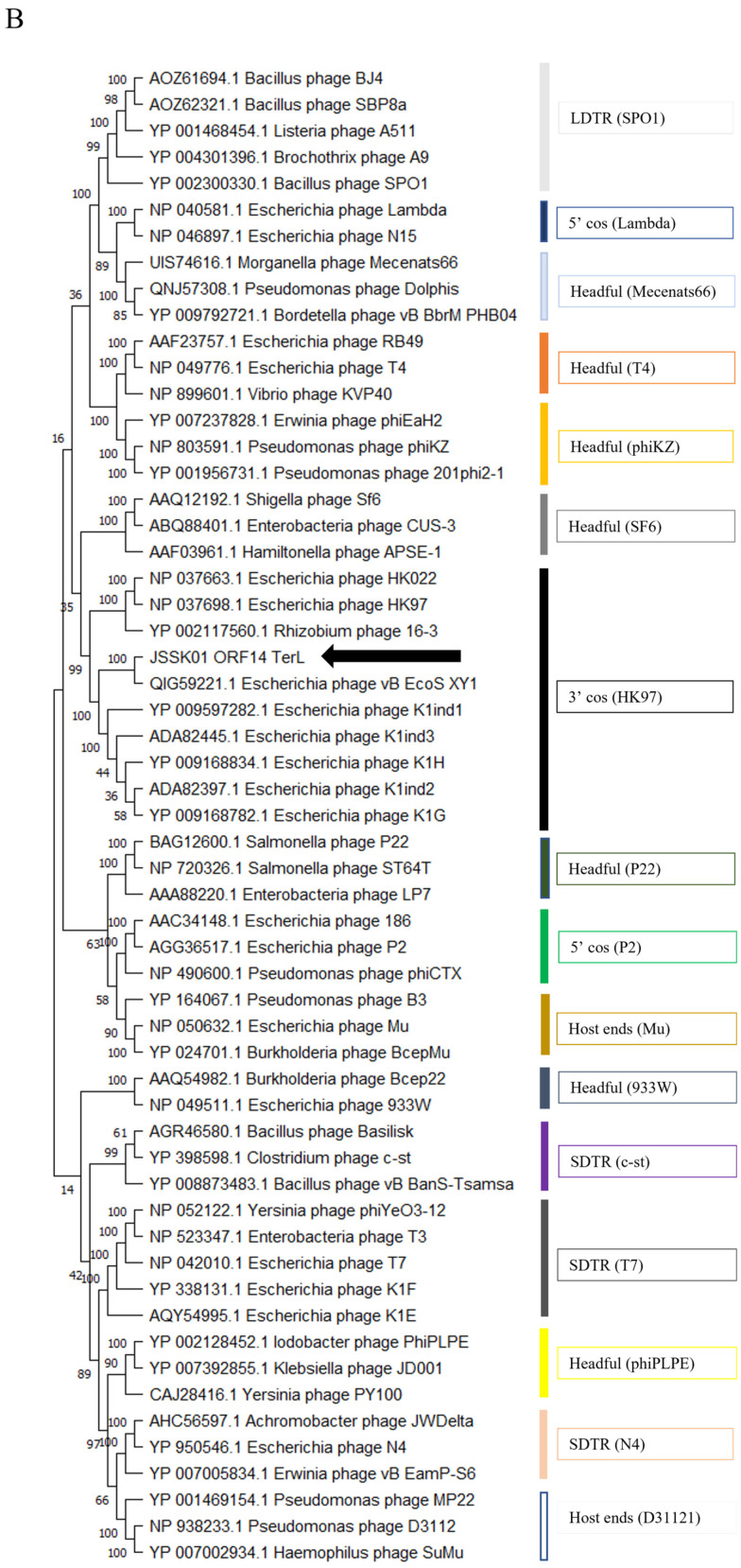
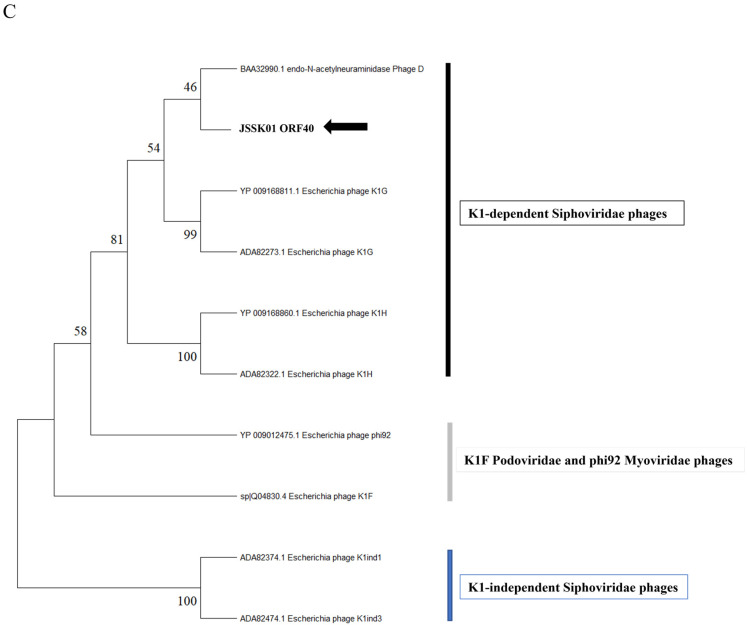
Figure 4.
Phylogenetic analysis of JSSK01. (A) JSSK01 phylogenetic analysis via VICTOR. Phage genome analysis for genome comparison with K1 gikeviruses, Jerseylikeviruses, and K1 podophages. The genomic distance was inferred via the D6 formula (scale length of 0.06). The relatedness was 2 at the family and subfamily levels, 3 at the genus level, and 21 at the species level. (B) Terminase large subunit-based phylogenetic analysis was carried out via MEGA 11 software. The JSSK01 terminase (ORF14) was compared with K1 gikevirus terminases and classified under 3’cos (HK97) terminases. (C) JSSK01 endo-N-acetylneuraminidase (ENgase) or ORF40 phylogenetic analysis was performed with other tail spike proteins via MEGA 11. The amino acid sequences of all K1 E. coli strains recognized by phage tail spike proteins were collected, and phylogenetic analysis was performed via the neighbor-joining method. The ORF40 of JSSK01 was grouped with K1-dependent tail spike proteins. For all the trees, the position of JSSK01 is marked with a black arrow.