Fig. 4 ∣. Expected results.
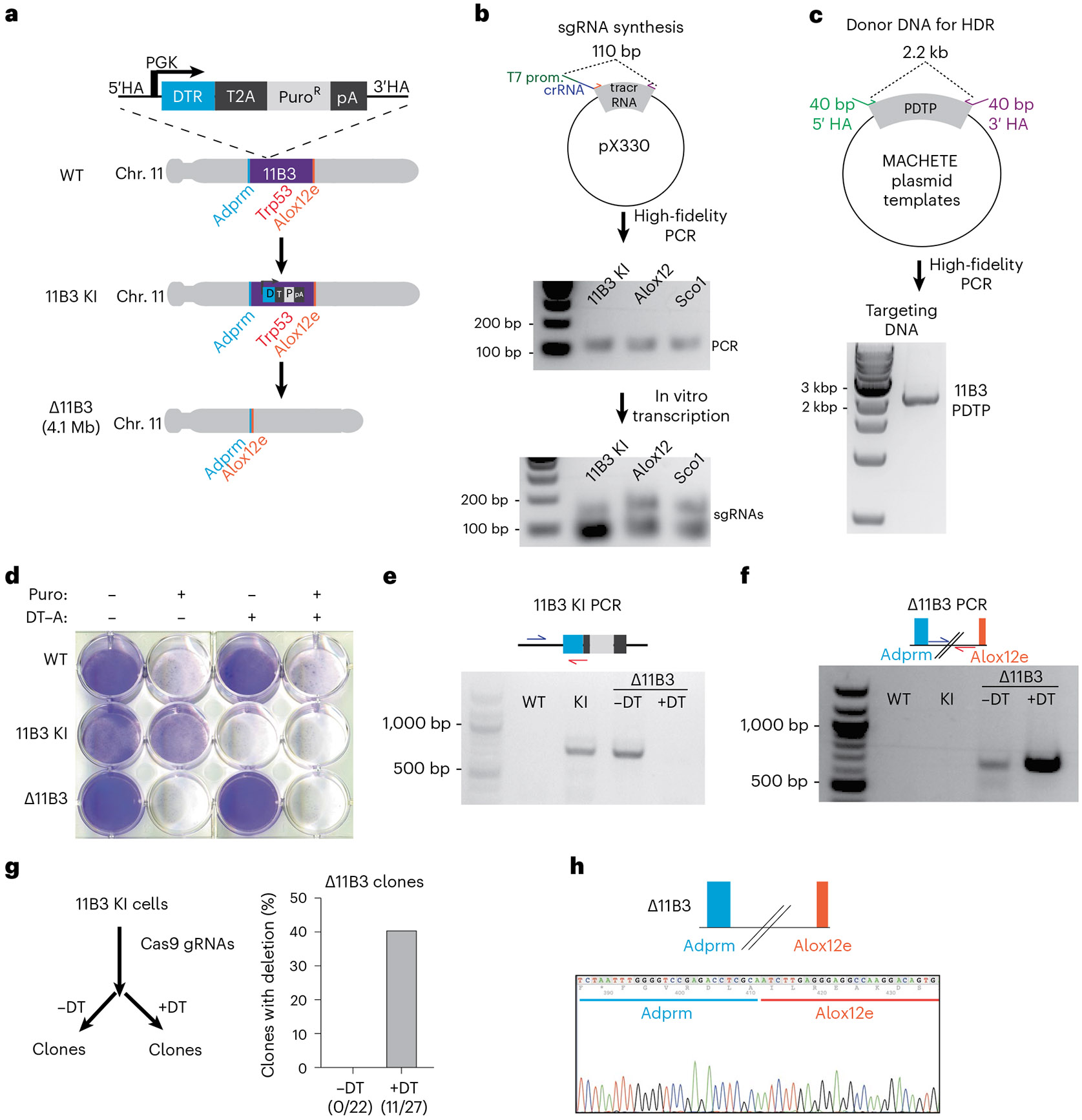
a, A schematic representation of the design to create a 4 Mb deletion of the mouse chromosome (Chr.) 11B3 locus containing the tumor suppressor Trp53 gene. b, A schematic of the procedure to synthesize KI and deletion sgRNAs for the murine 11B3 locus. Representative agarose gel of the PCR products used for in vitro transcription (top). Representative agarose gel of the transcribed sgRNAs, which can appear as doublets in these nondenaturing agarose gels (bottom). c, Schematic and representative images of the procedure to create the DNA donor for CRISPR-mediated HDR of a PGK DTR T2A Puro cassette into the 11B3 locus. A representative agarose gel of the DNA donor containing the MACHETE cassette and flanking homology arms used for HDR. d, Crystal violet staining of parental (WT), KI and deleted (Δ11B3) cells in the presence or absence of positive (puromycin, 1 μg/mL) and negative (diphtheria toxin, 50 ng/mL) selection. e, A PCR of the integration of the MACHETE cassette in the 11B3 locus. This was tested in the following cells: parental (WT), KI, unselected deleted (Δ11B3 − DT–A), and selected deleted (Δ11B3 + DT–A). f, A PCR of the engineered breakpoint of the 4.1 Mb Δ11B3 deletion. This was tested in the following cells: parental (WT), KI, unselected deleted (Δ11B3 − DT–A), and selected deleted (Δ11B3 + DT–A). g, Frequency of the intended Δ11B3 deletion in the population of unselected (− DT-A) and negatively selected (+ DT–A) cells. h, Representative Sanger sequencing of the Δ11B3 breakpoint PCR confirming the intended junction in genomic DNA. Images b–h reproduced from ref. 14, Springer Nature Ltd.
