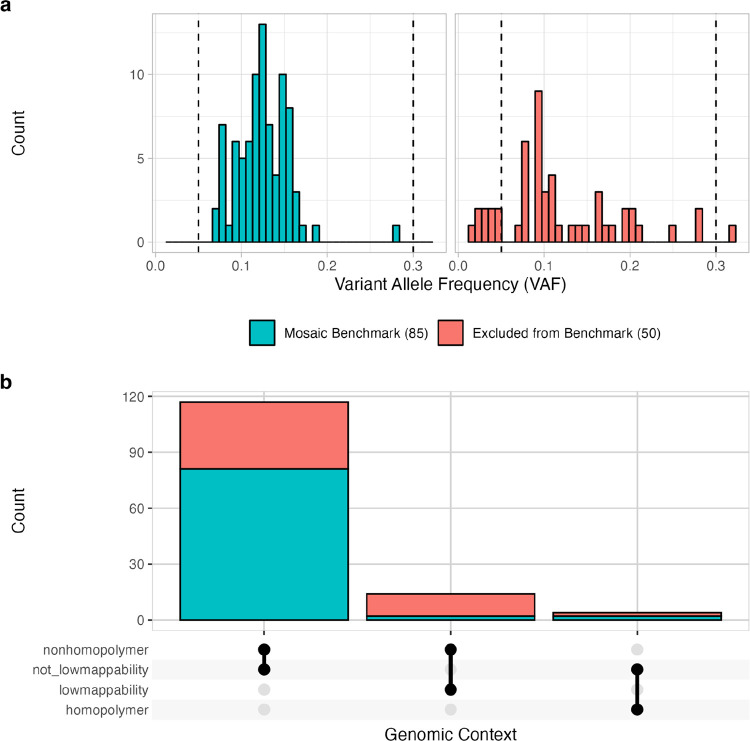
Figure 3.
SNV variant allele fractions (VAF) (a) for HG002-GRCh38 mosaic benchmark v1.0 and manually curated variants excluded from the benchmark. Values represent VAFs combined across all orthogonal technologies (BGI, Element, Illumina, PacBio Revio, and Sequel). Dashed vertical lines represent the targeted VAF range (5% - 30%) for the HG002 mosaic benchmark. Manually curated variant counts based on GIAB GRCh38 genome stratifications (b) reveal most mosaic benchmark v1.0 variants occur in easy-to-map and non-homopolymer regions of the genome.