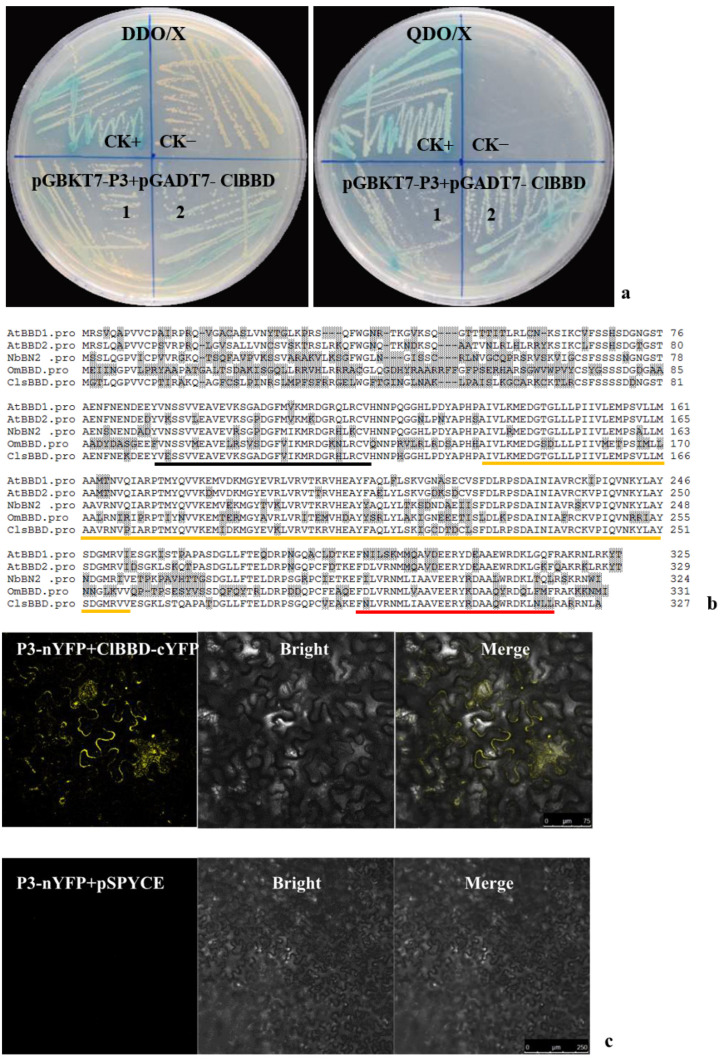
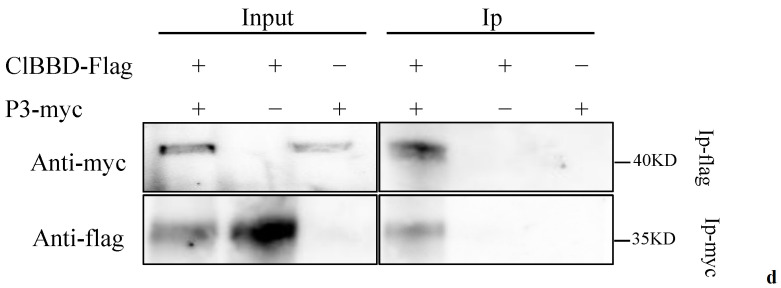
Figure 1.
The interaction of ZYMV P3 with ClBBD of watermelon. (a) Yeast cells containing pGBKT7-P3 were co-transformed with pGADT7-ClBBD on DDO/X (SD/-Leu/-Trp/X-α-Gal) and QDO/X (SD/-Leu/-Trp/-His/-Ade /x-α-gal) mediums. CK+, positive control for pGADT7-T + pGBKT7-53; CK−, negative control for pGADT7-T + pGBKT7-Lam. (b) Alignment of the deduced amino acid sequences of ClBBD with other plant homologs using the Clustal W algorithm. Different amino acids are highlighted in gray. Three domains, a highly conserved region (HCR) domain, an unknown function 151 (DUF151) domain, and a UV-responsive (UVR) domain, are underlined with black, orange, and red colors, respectively. (c,d) Verification of the interaction between P3 and ClBBD via bimolecular fluorescence complementation assays in N. benthamiana and co-immunoprecipitation (Co-IP) assays. YFP fluorescence was detected in the N. benthamiana leaves agroinfiltrated with pSPYNE-P3/pSPYCE-ClBBD using pSPYNE-P3/pSPYCE as negative control (c). Myc-tagged ZYMV P3 was co-expressed with Flag-tagged ClBBD in N. benthamiana. Proteins were immunoprecipitated (Ip) from total extracts using anti-myc or anti-flag antibodies and followed by Western blot using tag-specific antibodies (d).