Figure 1. VIR-7229 is a potent, pan-sarbecovirus neutralizing mAb.
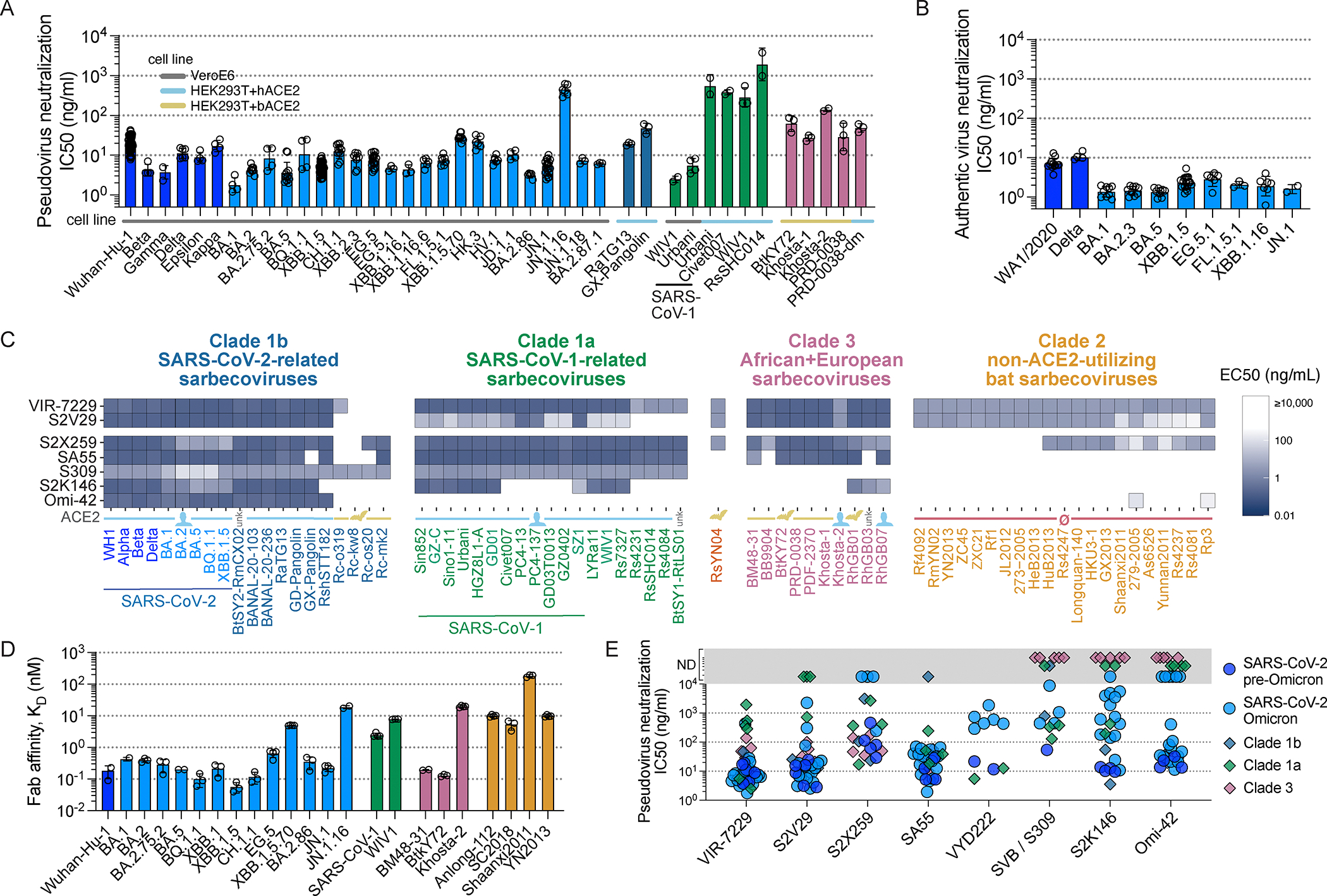
(A) VIR-7229-mediated neutralization of pseudoviruses. Bar color denotes sarbecovirus clade, as in panel C. Horizontal lines denote cell line employed in neutralization assay: VeroE6 (gray), HEK-293T+human ACE2 (cyan), HEK-293T+bat (R. alcyone) ACE2 (yellow). PRD0038-dm refers to PRD0038 harboring the K482Y/T487W RBD mutations (SARS-CoV-2 numbering 493Y/498W), which allow for entry using human ACE2.39,40 SARS-CoV-1 Urbani and WIV1 experiments were run in two assay conditions. See also Data S1. See Figure S2 for neutralization mechanisms of action.
(B) VIR-7229-mediated neutralization of authentic SARS-CoV-2 virus, performed with VeroE6 cells. The WA1/2020 isolate has the same S haplotype as Wuhan-Hu-1. See also Data S1.
(C) Breadth of VIR-7229 and comparator mAbs binding to a yeast-displayed panel of sarbecovirus RBDs spanning the known phylogenetic diversity. Line below the graph, denoted by “ACE2,” indicates whether a sarbecovirus binds or enters cells via human ACE2 (blue), bat but not human ACE2 (yellow), no ACE2 (pink), or unknown (unk.). See Figure 4A for phylogenetic relationships and clade definitions. See Data S1 for full sequences, phylogeny, and alignment.
(D) VIR-7229 Fab fragment binding affinity measured by SPR. Bar color denotes sarbecovirus clade. SARS-CoV-1 is Urbani. See also Data S1.
(E) Overview of pseudovirus neutralization by comparator mAbs, colored by sarbecovirus clade. Data points within the gray bar represent neutralization not detected (ND), i.e. IC50 >10,000 ng/ml. See Figure S1 and Data S1 for data separated by strain.
