Figure 3. Structural basis for VIR-7229 pan-sarbecovirus neutralization.
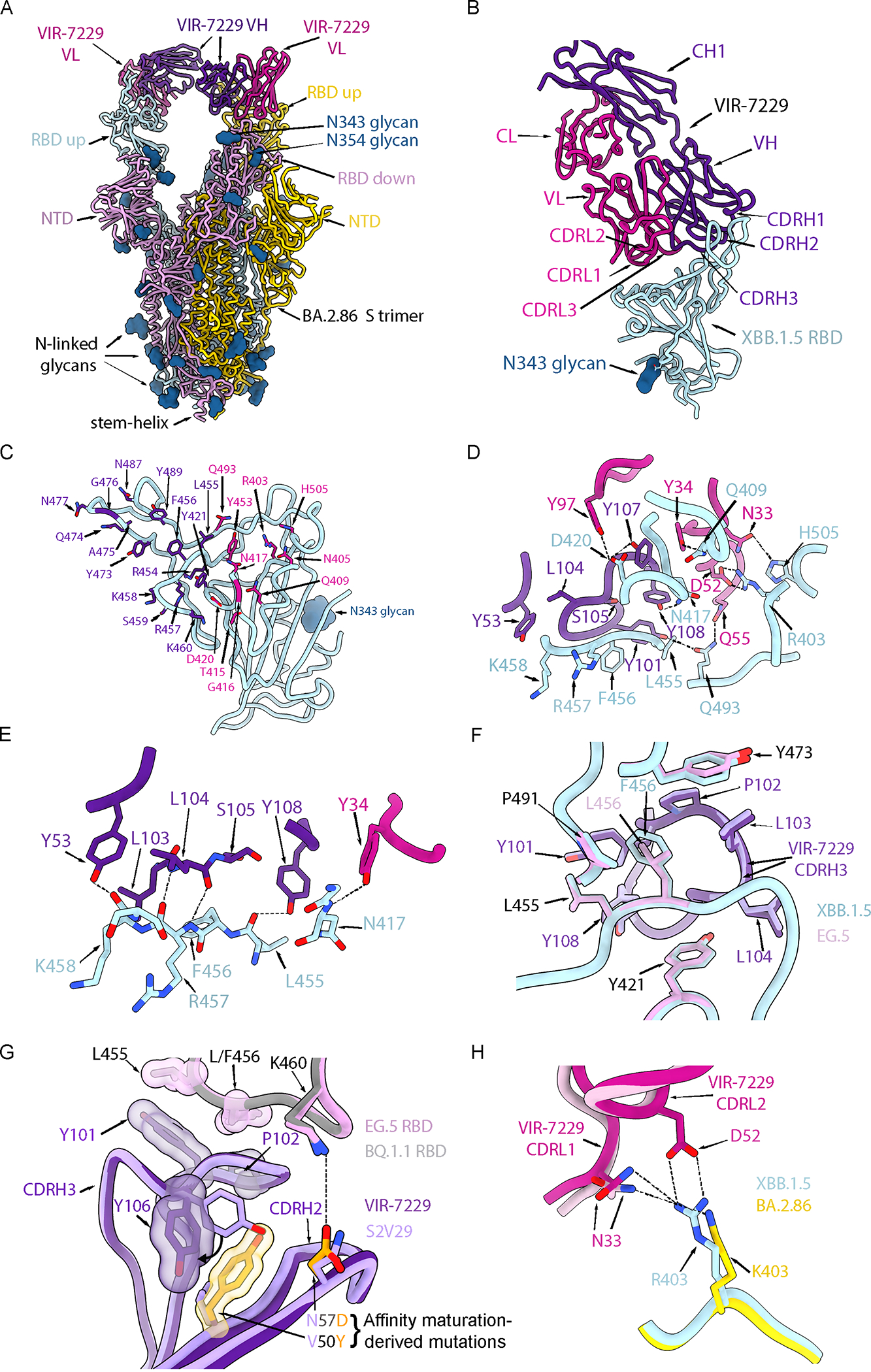
(A) Ribbon diagram of the cryoEM structure of the BA.2.86 S ectodomain trimer (cyan, pink and gold) in complex with two VIR-7229 Fabs (purple and magenta) with N-linked glycans rendered as blue surfaces. See also Figure S3.
(B) Ribbon diagram of the VIR-7229-bound XBB.1.5 RBD crystal structure. The bound S309 Fab is omitted for clarity. The N343 glycan is rendered as a blue surface. See also Figure S4.
(C) XBB.1.5 RBD (cyan) with VIR-7229 epitope residues shown as sticks and colored according to the (dominant) Fab interacting chain. RBD residues 420, 453, 455, 460 and 493 interact with the VIR-7229 heavy and light chains and were colored based on the chain with which they bury the greatest surface area.
(D) Zoomed-in view of select interactions formed between VIR-7229 and the XBB.1.5 RBD. Hydrogen bonds and salt bridges are indicated with black dash lines.
(E) Zoomed-in view of hydrogen-bonds (black dash lines) formed between VIR-7229 and the RBD backbone.
(F) Superposition of the VIR-7229-bound XBB.1.5 RBD (cyan RBD, dark purple mAb) and VIR-7229-bound EG.5 RBD (pink RBD, light purple mAb) showing accommodation of the F456L residue mutation.
(G) Superposition of the VIR-7229-bound EG.5 RBD (pink RBD, dark purple mAb) and S2V29-bound BQ.1.1 RBD (gray RBD, light purple mAb). The two CDRH3 residues differing between S2V29 and VIR-7229 (V50Y and N57D) are highlighted in orange. Select residues from the VIR-7229:EG.5 RBD structure are also shown as semi-transparent surfaces colored according to the sticks.
(H) Superposition of the VIR-7229-bound XBB.1.5 RBD (cyan RBD, dark pink mAb) and VIR-7229-bound BA.2.86 S (gold RBD, bright pink mAb) structures highlighting the conservation of electrostatic interactions (dashed lines) at the epitope/paratope interface due to the BA.2.86 R403K mutation. The D52 side chain is weakly resolved in the BA.2.86 S cryoEM density and was therefore not modeled.
