Figure 5. VIR-7229 has high tolerance for SARS-CoV-2 epitope variation.
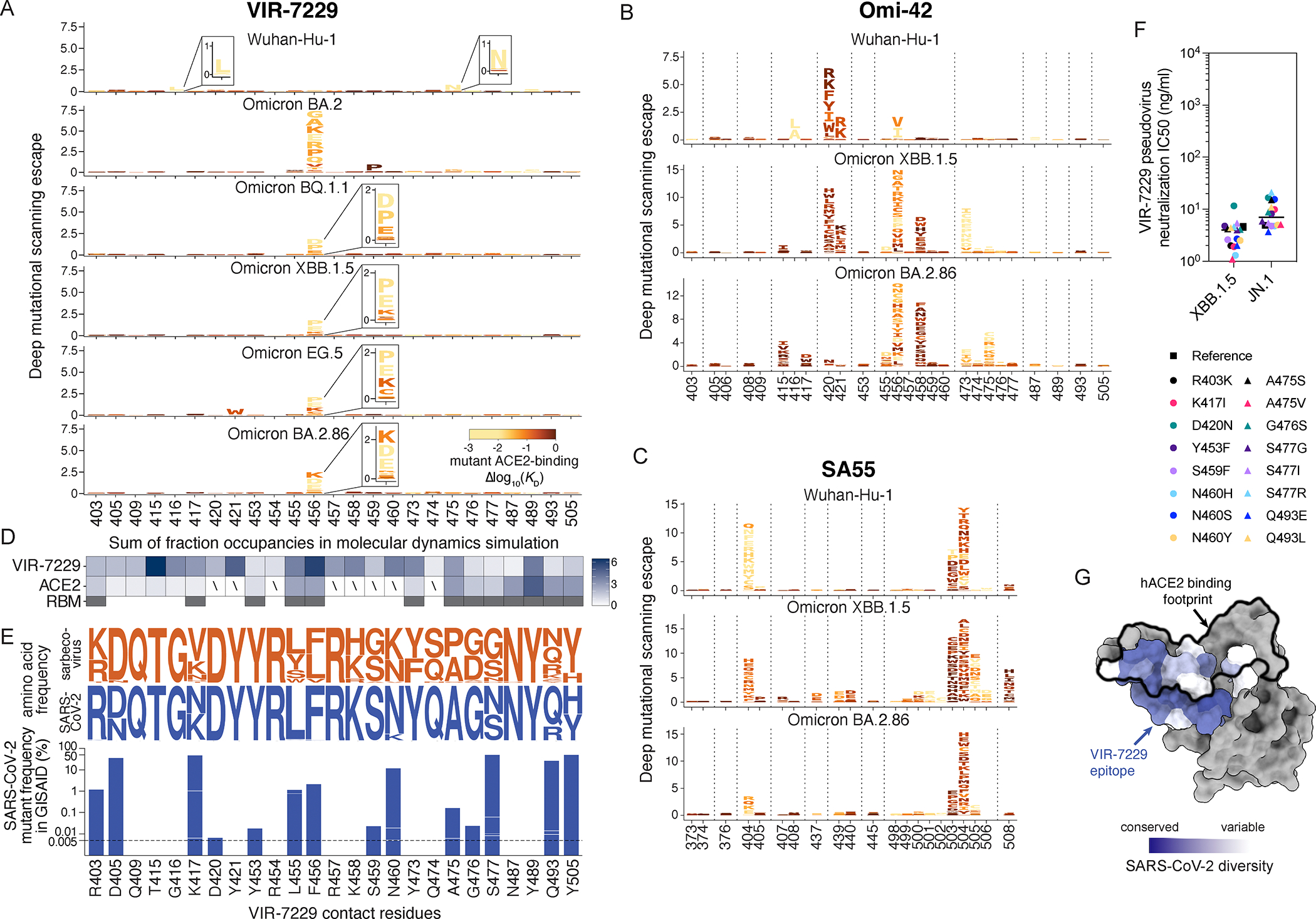
(A-C) Complete elucidation of mutations in the Wuhan-Hu-1, BA.2, BQ.1.1, XBB.1.5, EG.5, and BA.2.86 RBDs that enable escape from VIR-7229 (A), Omi-42 (B), or SA55 (C) binding using a yeast-display deep mutational scanning method. Letter height is proportional to mutant escape. Mutations are colored by their measured impacts on ACE2-binding affinity, where lighter yellow indicates increasingly deleterious effects on receptor binding (scale bar, bottom-right). See also Figures S5 and S6.
(D) Summary of ≥4.0-μs MD simulations of XBB.1.5 RBD bound to VIR-7229 or ACE2. Boxes are the number of persistent contacts at each RBD position in the VIR-7229 epitope, expressed as the fraction occupancy for each VIR-7229 or ACE2 contact across the MD simulation, added together for each RBD position. See panels A or E for RBD position annotations. Slash indicates no contact, i.e. sum of fraction occupancy <0.1. Full glycans were modeled into the RBD:ACE2 X-ray structure; some ACE2 contacts are glycan-mediated, see Figure S4F. The third row indicates RBM residues (gray boxes), defined as RBD:ACE2 protein:protein contacts within 5 Å in the X-ray structure. See also Figure S4 and Data S4.
(E) Top, logoplots illustrating the frequency of amino acid variation in VIR-7229 epitope residues across human-ACE2-utilizing sarbecovirus sequences (orange) and SARS-CoV-2 sequences available on GISAID from May 8, 2024 (blue). Bottom, barplot illustrating SARS-CoV-2 mutant frequencies (log scale; residues present in the ancestral Wuhan-Hu-1 sequence are not plotted) for all mutants with >0.005% occurrence in GISAID (up to May 8, 2024). VIR-7229 neutralization of each of these mutations was validated via neutralization of single mutants introduced into XBB.1.5 and JN.1 pseudovirus (panel F) or presence of a mutation in a circulating variant that is neutralized (Figure 1A), with the latter mutations labeled with asterisk.
(F) VIR-7229-mediated neutralization of SARS-CoV-2 epitope variants with >0.005% frequency in GISAID (panel E), tested on the XBB.1.5 and JN.1 backgrounds. “Reference” refers to XBB.1.5 or JN.1 with no additional amino acid substitutions. Substitutions are annotated relative to the Wuhan-Hu-1 sequence. R403K is part of the JN.1 reference sequence. See also Data S1.
(G) SARS-CoV-2 conservation of the VIR-7229 epitope mapped to SARS-CoV-2 Wuhan-Hu-1 RBD structure (PDB 6M0J). ACE2 binding footprint is illustrated as a black outline.
