Figure 6. VIR-7229 exhibits a high barrier to viral escape.
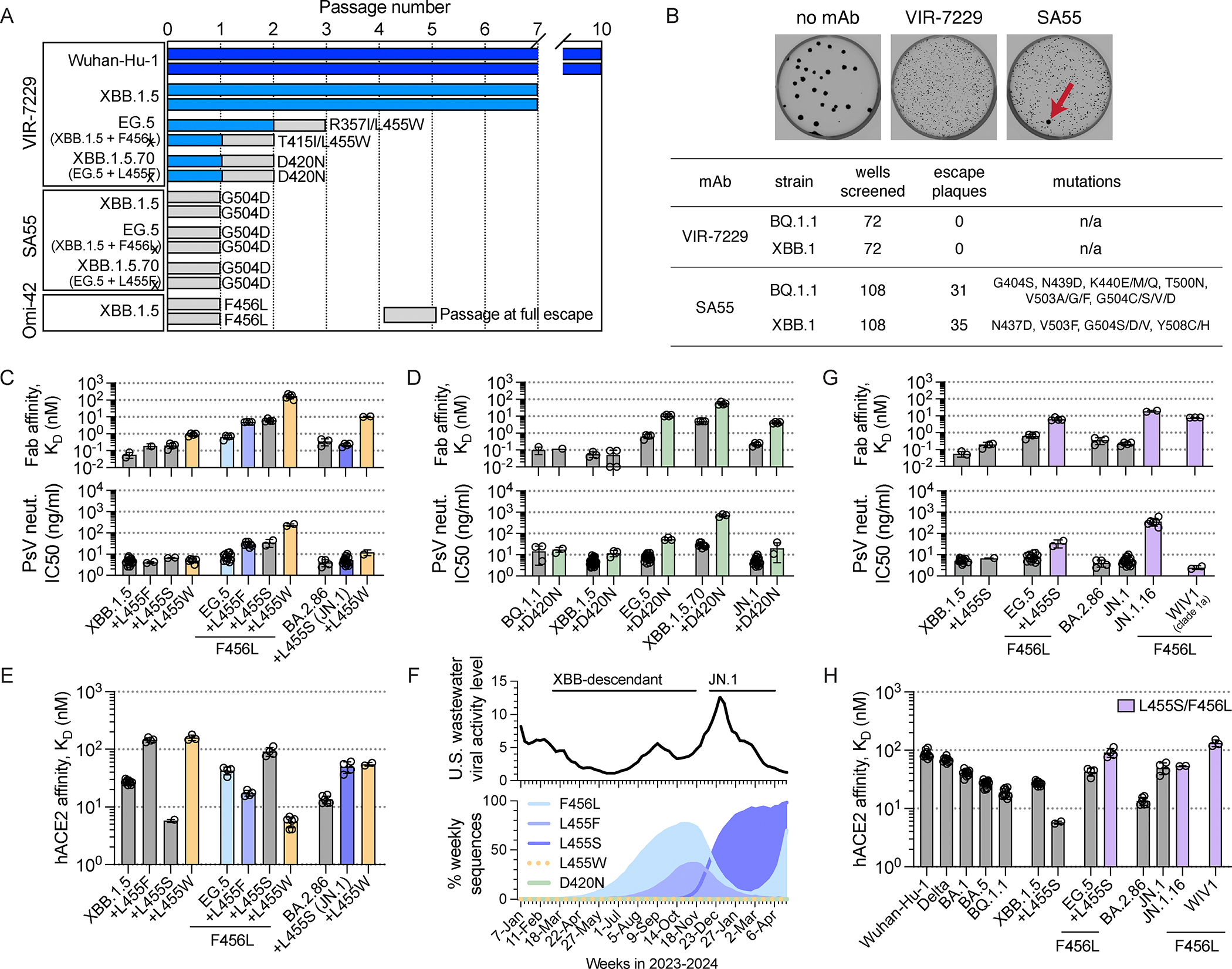
(A) Serial passaging of Wuhan-Hu-1 and XBB.1.5 rVSV in the presence of mAb did not result in escape from VIR-7229, as defined by ≥20% cytopathic effect in the presence of 20 μg/mL mAb (experiment terminated after 10 and 7 passages, respectively) whereas XBB.1.5 rVSV escaped from comparator mAbs (SA55 and Omi-42) after a single passage. EG.5 rVSV escaped from VIR-7229 after two to three passages, and from a comparator mAb (SA55) after a single passage. XBB.1.5.70 rVSV escaped from VIR-7229 after two passages and from a comparator mAb (SA55) after a single passage. Two independent replicates were performed for each experiment. Figure shows RBD mutations observed after sequencing; the T941K mutation was also observed in one replicate of the XBB.1.5.70 serial passaging with VIR-7229. See also Figure S6 and Data S5.
(B) Plaque-based selection of BQ.1.1 and XBB.1 rVSV escapes was performed with VIR-7229 and comparator mAb SA55. Zero escape plaques were observed in 72 independent selections for VIR-7229 whereas 31 and 35 escape plaques, respectively, were observed in 108 independent selections for SA55. Representative images from BQ.1.1 selection are shown, red arrow indicates escape plaque. See also Data S5.
(C) Impact of mutations at RBD position 455 on VIR-7229 Fab fragment binding affinity measured by SPR (top) and on VIR-7229-mediated pseudovirus neutralization (bottom). EG.5+L455F is XBB.1.5.70. Colored bars correspond to mutations plotted in panel F. See also Data S1.
(D) Impact of the D420N mutation on VIR-7229 Fab fragment binding affinity measured by SPR (top) and on VIR-7229-mediated pseudovirus neutralization (bottom). See also Data S1.
(E) Impact of mutations at RBD position 455 on ACE2 affinity measured by SPR. Colored bars as in panel C. See also Data S1.
(F) Top – SARS-CoV-2 viral activity level in U.S. wastewater, January 2023 – April 2024 (cdc.gov). Bottom – Frequency of SARS-CoV-2 S mutations as percentage of weekly sequences deposited in GISAID, January 2023 – April 2024. L455W and D420N frequencies are too low to be visible. As of May 8, 2024, >96% of L455F and >86% of L455W mutations co-occur with F456L, primarily in EG.5 and derivative strains; approximately 94% of L455S mutations are in a BA.2.86/JN.1 background.
(G) Impact of L455S +/− F456L on VIR-7229 Fab fragment binding affinity measured by SPR (top) and on VIR-7229-mediated pseudovirus neutralization (bottom). See also Data S1.
(H) Impact of L455S +/− F456L mutations on ACE2 affinity measured by SPR. See also Data S1.
