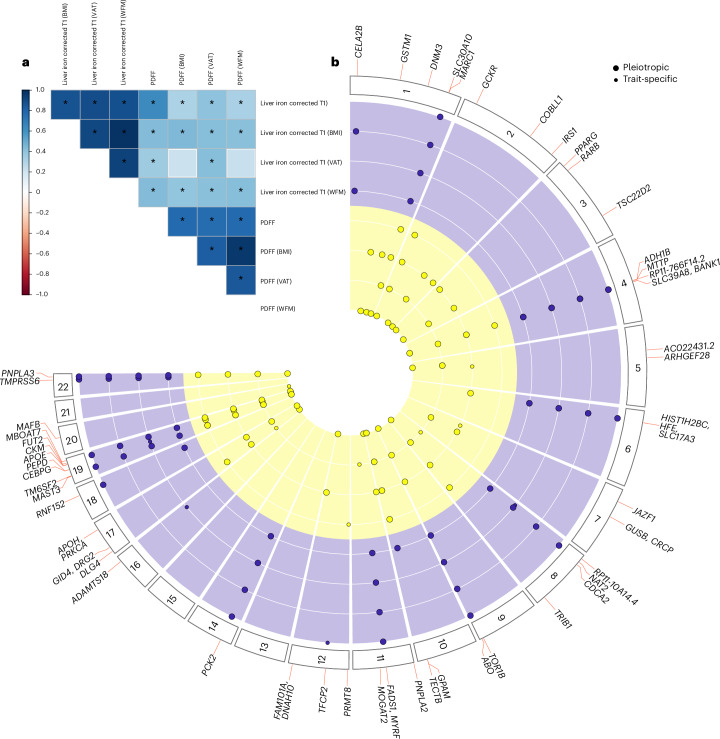
Fig. 1. Overview of the identified loci for liver triglycerides and inflammation/fibrosis by the multi-adiposity-adjustment GWAS.
a, Genetic correlation among different multi-adiposity-adjusted PDFF and liver iron corrected T1 was estimated using LD score regression analysis. The asterisks denote Benjamini–Hochberg false discovery rate (FDR) <0.05. The color bar represents the genetic correlation values. Detailed summary statistics for genetic correlations have been reported in Supplementary Table 3. b, Circular Manhattan plot of PDFF and liver iron corrected T1 for different adiposity adjustments. The association analyses were performed using REGENIE adjusting for adiposity index, age, sex, age × sex, age2 and age2 × sex, first ten genomic principal components and array batch. Each dot represents an independent genetic locus. Yellow represents loci associated with liver PDFF and purple represents those associated with liver cT1. Large dots represent pleiotropic loci (where the association with either PDFF or liver cT1 was shared among two or more adiposity adjustments). Small dots show adiposity-trait specific associations. Loci in bold are shared among both traits irrespective of the adiposity adjustment. Only loci with a genome-wide significant P <5 × 10−8 calculated by a whole-genome regression model (Methods) are shown. P values were two-sided and not corrected for multiple testing among four different models (unadjusted, adjusted for BMI, WFM and VAT).