Figure 3.
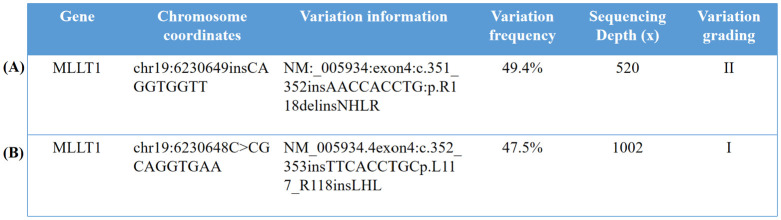
Identification and characterization of MLLT1 variants. (A) The whole genome chip detection of the solid tumor sample of the 6-month-old male infant showed no loss of heterozygosity (LOH) variation in the 1p and 16q regions, no repetitive variation in the 1q region, and no other acquired chromosome copy number variation (CNV) changes. (B) The results of the whole genome chip detection sample of the 9-month-old male infant were 0 acquired CNV abnormalities, no LOH variation found in the 1p region and the 16q region, and no acquired variation was found in the 1q region. (Variation grading in the table is divided into grade I, variation clearly related to the disease; level II, variations that may be associated with the disease; and grade III, variants with unknown clinical significance.). 1. In our study, the reference genome of the chromosome location was GHCh37/hg19. 2. In our study, the detection method used for the two children with nephroblastoma was whole-genome sequencing. The whole genome chip belongs to Illumina cytoSNP-850K chip. The basic principle is to connect the probe to the microbeads, and then, the microbeads carrying the probe are randomly adhered to the chip. The DNA of the sample to be tested is hybridized with the probe and the single base extension is performed. The copy number variation (CNV) of the sample to be tested is analyzed by scanning the fluorescence signal. The probe position can cover the whole genome of 22 pairs of autosomes and sex chromosomes and can detect chromosome copy number abnormalities (deletion or duplication) and uniparental disomy (UPD) with a normal copy number.