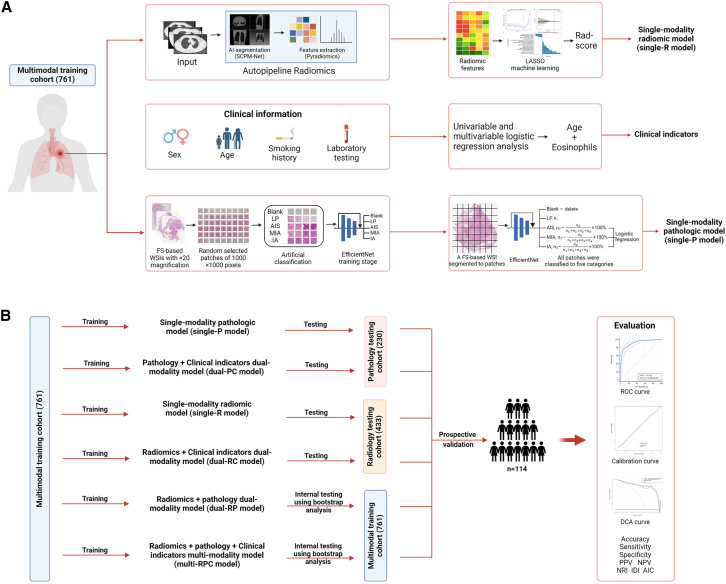
Figure 2.
Workflow of the study
(A) Images of non-enhanced thoracic CT and FS-based hematoxylin and eosin-stained WSIs with 20× magnification were collected and segmented for radiomic feature extraction and pathologic parameter generation, respectively. The classification of five pathologic categories (Blank, LP, AIS, MIS, and IA) for WSI patches was accomplished through pretrained EfficientNet model. Single-R model was developed with Rad-score, and single-P model was developed with AIS%, MIA%, and IA%. Clinical indicators, including age and eosinophils, were selected by using univariable and multivariable logistic regression analysis. All dual-/multimodality models were developed by logistic regression.
(B) All models were trained in the multimodality training cohort, while single-P and dual-PC models were tested in the pathology testing cohort, single-R and dual-RC models were tested in the radiology testing cohort, and dual-RP and multi-RPC models underwent internal testing by bootstrap analysis in the multimodality training cohort. All models were further validated in the prospective validation cohort. FS: frozen section; WSI: whole-slide image; LP: lung parenchyma; AIS, adenocarcinoma in situ; MIA, minimally invasive adenocarcinoma; IA, invasive adenocarcinoma. AUC, area under the curve; NPV, negative predictive value; PPV, positive predictive value; NRI, net reclassification improvement; IDI, integrated discrimination improvement; AIC, Akaike information criterion.