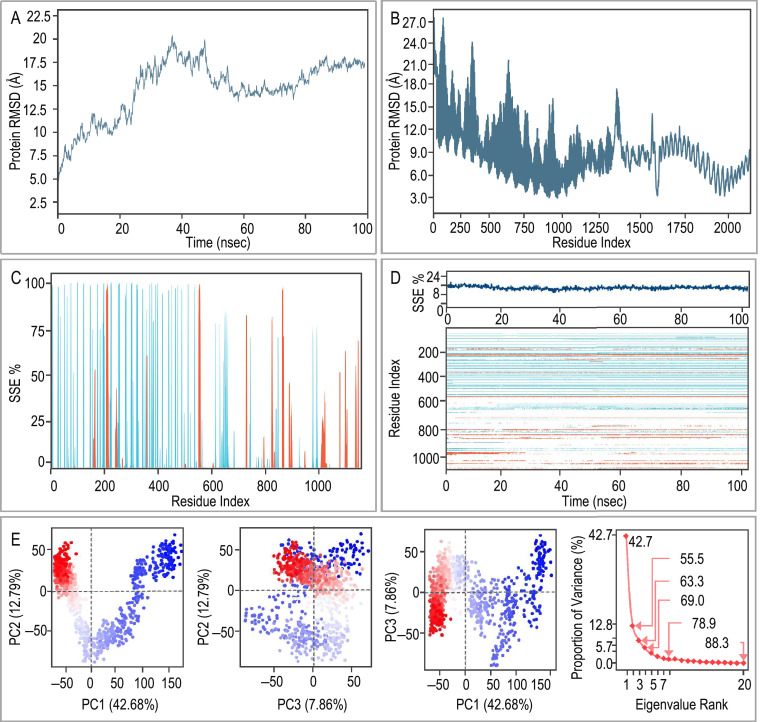
Fig. 5.
MD simulations and principal component analysis (PCA) of the ZL12138L vaccine-TLR-4 complex. (A) Root Mean Square Deviation (RMSD) plot of the ZL12138L vaccine-TLR-4 complex over 100 ns of MD simulation. The RMSD values indicate structural deviations from the initial configuration, providing insights into the stability of the complex over time. (B) RMSD fluctuation per residue index over the simulation time. This plot highlights specific regions within the ZL12138L vaccine-TLR-4 complex that exhibit appreciable conformational changes, suggesting potential flexible and stable regions. (C) Secondary structure element (SSE) distribution along the residue index. The plot indicates the percentage of time each residue spends in specific secondary structures (e.g., α-helices and β-strands) throughout the simulation, providing insights into the structural dynamics of the complex. (D) Time evolution of the SSE. The upper part of the plot shows the percentage of each SSE over time, and the heatmap below illustrates the temporal changes in secondary structure along the residue index, highlighting regions of structural transitions and stability during the simulation. (E) PCA of the ZL12138L vaccine-TLR-4 complex. The first panel shows a projection of the motion along the first two principal components (PC1 and PC2), capturing 42.68% and 12.79% of the variance, respectively. The clusters in the plot indicate distinct conformational states visited during the simulation. The second panel shows the projection of the motion along PC2 and PC3, capturing 12.79% and 7.86% of the variance, respectively. The third panel shows the projection of the motion along PC1 and PC3. The last panel shows the plot of the proportion of variance explained by each principal component, with an inset showing the cumulative variance. This analysis quantifies the contribution of each mode to the overall motion, demonstrating the dominant dynamics captured by the first few principal components.