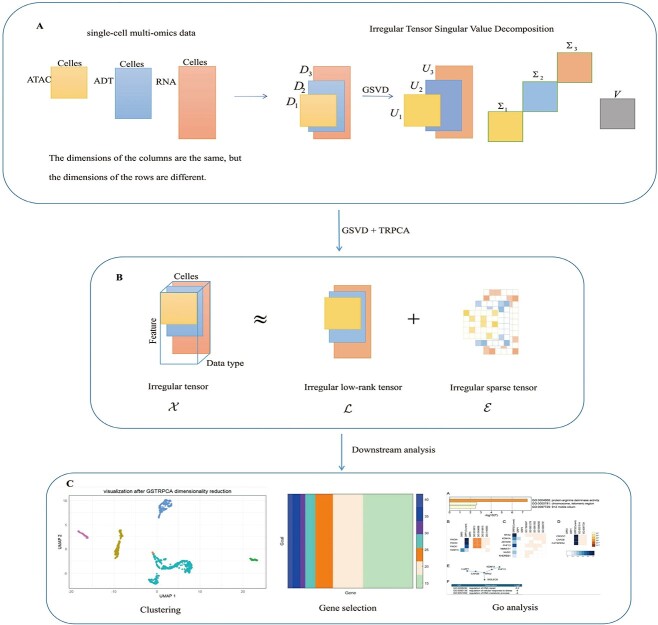
Figure 1.
Comprehensive overview of GSTRPCA. (A) Input single-cell multi-omics data types and construct them as irregular tensor data for the generalized singular value decomposition of irregular tensors. (B) Decompose the irregular tensor data into an irregular low-rank tensor and a sparse tensor, which is more complex than a straightforward low-rank approximation, as it incorporates considerations of sparsity. In the depicted irregular sparse tensor, the majority of the elements are zero. (C) We conducted a comprehensive analysis of single-cell multi-omics data, employing UMAP (uniform manifold approximation and projection) for the visualization of cellular clustering. Additionally, we performed gene selection and GO enrichment analysis to identify biological processes and functions associated with the selected genes.