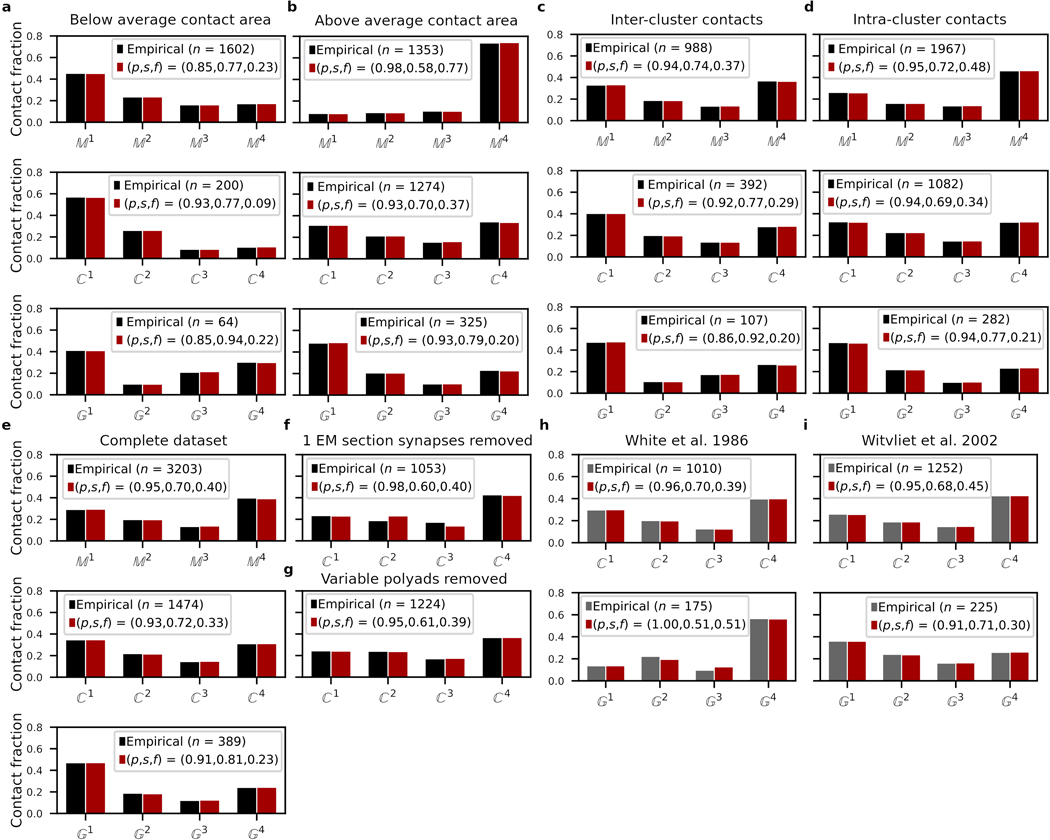
Extended Data Fig. 3. Core and variable model validations.
a,b, Model fits for reproducibility of contacts, with membrane contact areas a, below and b, above the log-normalized mean (after thresholding, Methods, Extended Data Fig. 2h). c,d, Reproducibility model fits of c, inter-cluster and d, intra-cluster contacts. e, Reproducibility model fits for complete , and datasets including membrane contact areas <35% (results qualitatively similar to restricted dataset model fit in Figure 2a; Methods: Generating reference graphs). f, Reproducibility model fits for excluding synaptic contacts scored in only 1 EM section (Methods). g, Reproducibility model fits for excluding synaptic contacts derived from non-reproducible post-synaptic partners of polyadic synapses (Methods). h,i, Reproducibility model fits for synaptic and gap junction contact datasets scored by h, White et al. (1986)5 and i, Witvliet et al. (2020)20 limited to our contacts. : membrane, : chemical synapse and : gap junction contacts. Black bars: empirical distributions used in this study. Gray bars: other empirical distributions5,20. Red bars: Model fits for the empirical distributions. All fractions of the total empirical counts ().