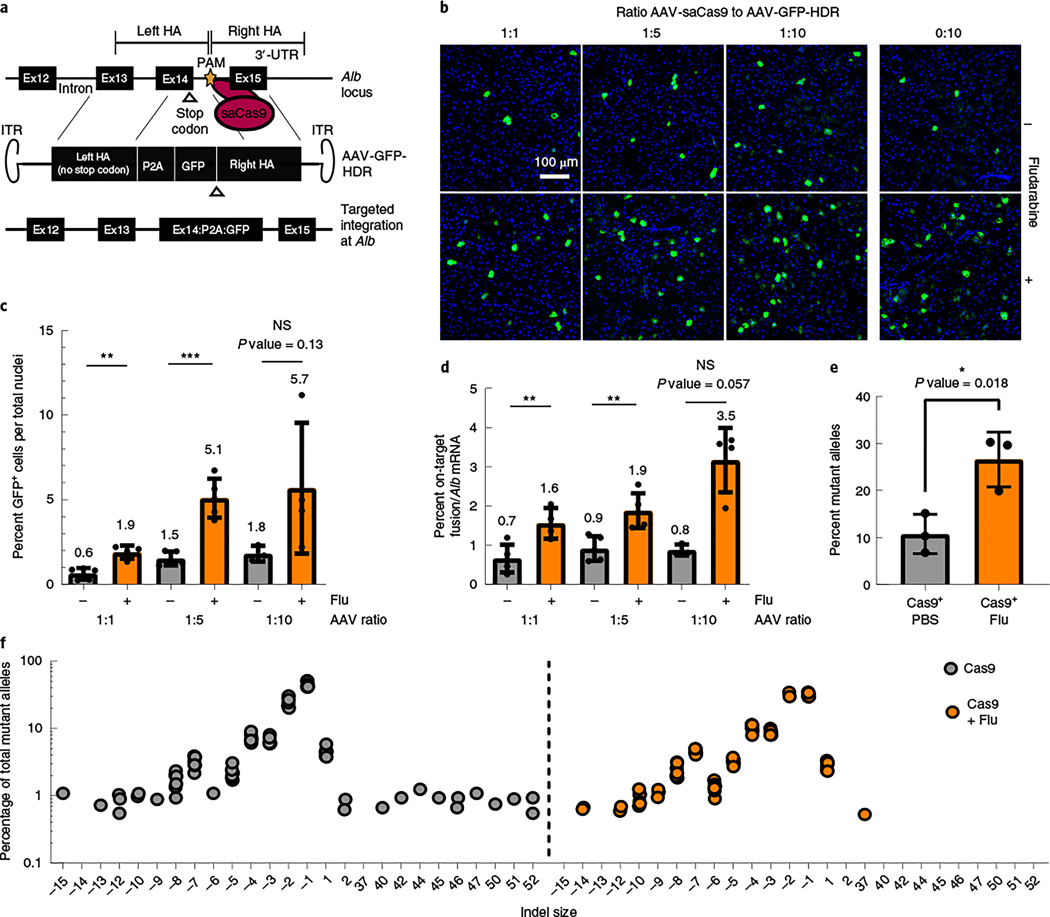
Fig. 5 |. Fludarabine increases CRISPR/Cas9 gene editing efficiency in vivo.
a, saCas9 is targeted to the intron downstream of exon 14 in the mouse Alb locus, delivered by AAV-saCas9. Homology-directed repair is accomplished using an AAV-encoded repair template, AAV-GFP-HDR, which contains a P2A-GFP transgene flanked by sequences homologous to the gRNA target site. The PAM site of AAV-GFP-HDR was mutated to avoid self-targeting; 3′-UTR, 3′-untranslated region. ITR, inverted terminal repeats. b, Mice were treated with PBS or fludarabine, as previously described, and co-injected with AAV-saCas9 and AAV-GFP-HDR. Three ratios of AAV-saCas9 to AAV-GFP-HDR vector were used (1:1, 1:5 and 1:10), with each part representing 6.0 × 1012 viral genomes per kilogram of body weight. Two weeks later, mice were sacrificed, and livers were imaged for native GFP expression. Representative images are shown; n = 3 to 5 mice per group. c, GFP+ cells were quantified from multiple images per mouse. A Shapiro–Wilk test was used to test for normal distribution, an F-test determined variation between groups, and a two-tailed Student’s t-test or t-test with Welch’s correction tested for significance. Each point represents data from an individual mouse; n = 3 to 5 mice per group. Bars represent the group mean, with error bars representing s.d. The P value was 0.0012 for the 1:1 group, 0.0002 for the 1:5 group and 0.138 for the 1:10 group. d, Total RNA was extracted from mouse livers and used for quantification of the on-target HR-derived fusion mRNA or Alb mRNA by qPCR. mRNA levels were normalized to Actb mRNA. Data are displayed as a percentage of fusion mRNA out of all Alb mRNA. Each point represents data from an individual mouse; n = 3 to 5 mice per group. Bars represent the group mean, with error bars representing s.d. Significance testing was performed as in c. The P value was 0.0084 for the 1:1 group, 0.004 for the 1:5 group and 0.057 for the 1:10 group. e, Separately, mice were injected with 6.0 × 1012 viral AAV-saCas9 genomes per kilgram of body weight with or without fludarabine treatment. Two weeks later, gDNA was extracted from livers, and targeted deep sequencing of the gRNA target site was performed. Data are normalized to sequencing data from a non-injected control mouse and are displayed as a percentage of alleles containing indels; n = 3 mice per group. Bars represent the group mean, and error bars represent s.d. Significance testing was performed with a two-tailed t-test after testing for distribution and variance. f, The top 12 mutant alleles from targeted deep sequencing were analyzed for all mice and technical replicates. Indels were graphed as the percentage of reads from all mutant alleles based on size (x axis).