Fig. 5. |. Expression vectors and construct generation.
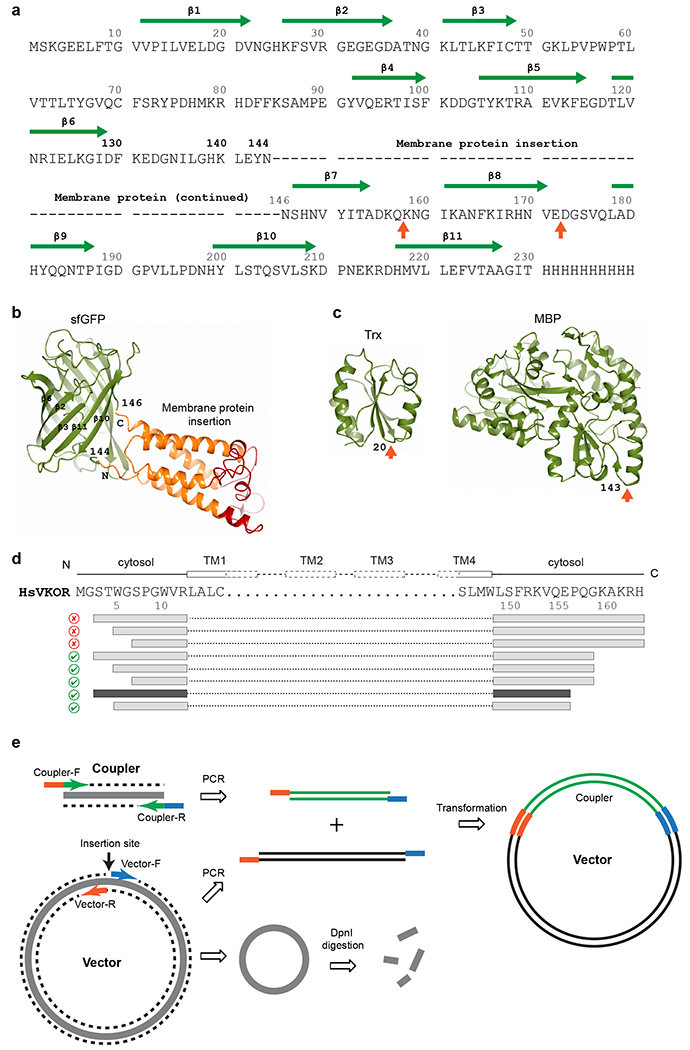
a, The sequence of sfGFP and insertion sites for a membrane protein. The first choice of insertion (dashes represent membrane protein sequence) is between Asn144 and Asn146 in sfGFP. Other potential insertion sites55,56 are indicated by orange arrows below the sequence. The β-strands and residue numbers of sfGFP are indicated above the sequence. The C-terminal 8 residues (231-238) of sfGFP have been removed to reduce its structural flexibility and to favor stable crystal packing. b, Structure of sfGFP (shown in green) with fused human VKOR (transmembrane region in orange and extramembrane region in red). The Asn144-Asn146 insertion site is indicated. c, Structures of other couplers, thioredoxin (Trx) and maltose binding protein (MBP), that we have tested. Insertion sites are indicated by orange arrows. d, Serial truncations exemplified by human VKOR constructs. The FSEC results are indicated by good (green checkmark) and bad (red cross) icons. Interestingly, truncations at the C-terminus (removing the ER retention signal, KAKRH) rescue the expression of restrained constructs. Further truncations result in the final construct (dark grey bars) that is used to obtain the high-resolution structure. e, Simplified seamless cloning38. The coupler coding sequence and the entire expression vector are PCR amplified. The primers for coupler contain sequences (orange and blue) overlapping with the primers for the vector. DpnI digestion removes the original methylated template. The two PCR fragments can assemble to form the coupler-inserted plasmid with considerable efficiency after transformation into DH5α E. coli cells.
