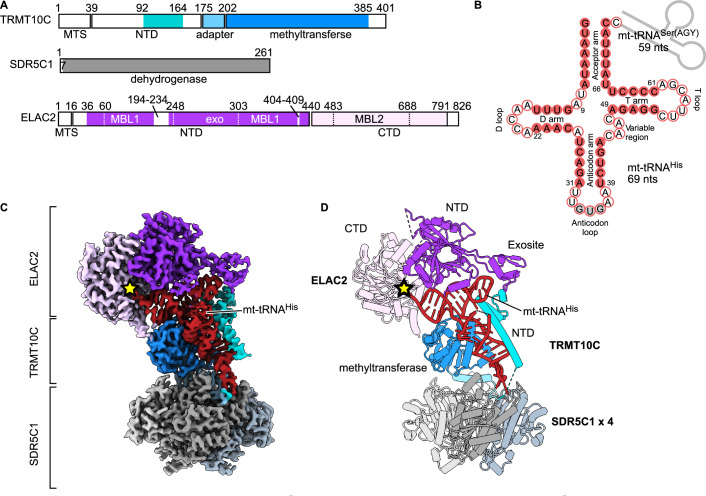
Figure 1. Structure of human mitochondrial RNase Z.
(A) Domain architecture of the human mitochondrial RNase Z components. The N-terminal (NTD) and C-terminal (CTD) domains of ELAC2 are shown in dark and light purple, respectively. MBL1 and MBL2, which are the metallo-β-lactamase folds, and the exosite insertion are indicated with dotted lines. TRMT10C is shown in blue, with the NTD highlighted in cyan. MTS, mitochondrial targeting sequence. Unmodelled regions are in white. (B) The mt-tRNA precursor consists of tRNAHis (red) and tRNASer(AGY) (gray line), of which the latter has been cleaved off in the ternary product structure. Base-paired nucleotides are in red circles, and non-base-paired ones are in white circles. The anticodon triplet is in gray circles. Canonical tRNA numbering is indicated (Giegé et al, 2012) and is used throughout the text (the correspondence with nucleotide numbers in mitochondrial tRNAHis, which was used in the PDB deposited structures, is shown in Appendix Table S1). (C, D) Composite cryo-EM density (C) and cartoon representation (D) of the RNase Z complex. Colored as in A. The four SDR5C1 subunits are shown in different shades of gray. The ELAC2 active site is marked with a yellow star.