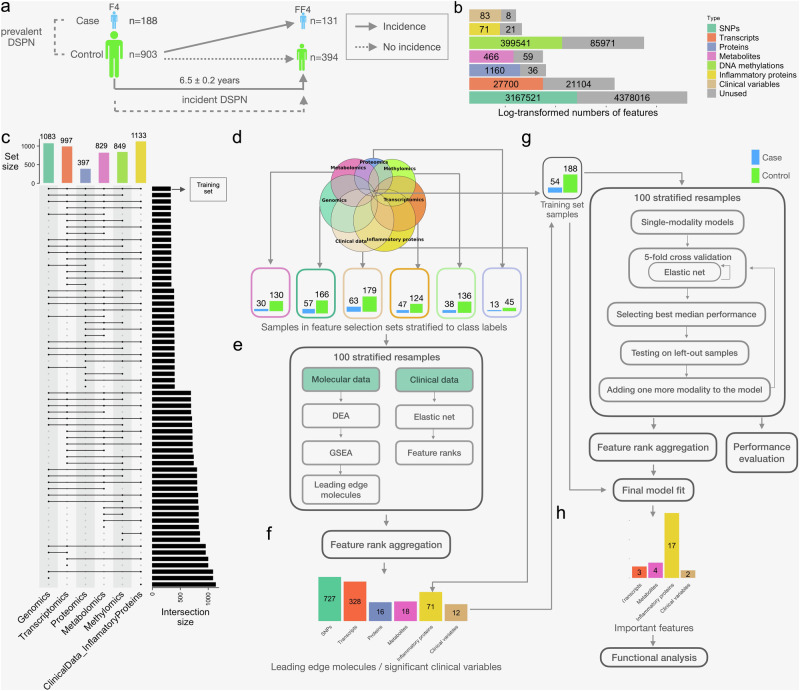
Fig. 1. Workflow of interpretable multimodal framework for feature prioritisation, DSPN classification and disease incidence prediction.
a Distribution of samples across time points (KORA F4 and FF4), disease status (case or control) at baseline (KORA F4) and follow-up (KORA FF4) and prediction tasks. Both models were trained on the same set of F4 features but different labels and a subset of samples. b Number of features stratified according to data modalities. In grey are removed features after pre-processing. c Number of samples characterised within each data modality and their overlaps in KORA F4. d Fully characterised samples in KORA F4 were exclusively leveraged for g the second and final training step, whilst the remaining sparse samples were used for e prior feature prioritisation: All molecular features were shortlisted based on differential expression analysis (DEA), gene set enrichment analysis (GSEA) and their leading-edge genes (“Methods”), whilst clinical features were ranked according to feature importance of elastic net models. f Features for the final training step were selected based on rank aggregation (“Methods”). g The final training set contained 54 DSPN cases and 188 controls in KORA F4. In the second step, elastic net models determined the optimal number of modalities, features and combination of modalities. These models implemented forward feature selection in a nested cross-validation, using weighted log loss to account for class imbalance, and finally 100 stratified resampling during training and rank aggregation (“Methods”), thus returning h the refined and final model further subject to functional analysis for gaining insights in DSPN pathophysiology.