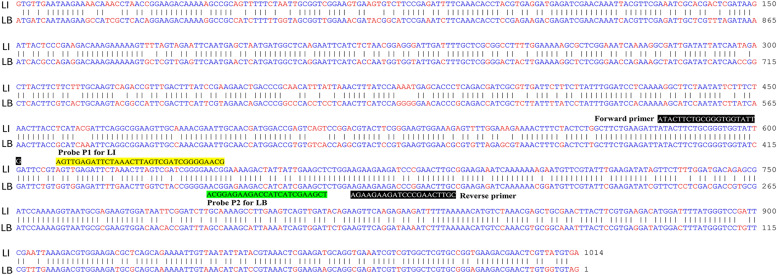
Fig. 3.
Sequence alignment for each primer/probe pair using representative Leptospira species. Alignment of the DNA sequence of the fliG gene from L. interrogans serovar Copenhageni str. Fiocruz L1–130 (NCBI accession number: NC_005823.1) and the fliG2 gene from L. biflexa serovar Patoc strain 'Patoc 1 (Paris)' (NCBI accession number: CP000786.1) was performed. Alignment of the DNA sequence using the Martinez/Needlemen-Wunsch alignment method by MegAlign (DNASTAR, USA) revealed that the sequence similarity index was 78.3 %. Identical nucleotides are shown as blue letters, and different nucleotides aligned at the same position are shown as red letters. The positions of the forward and reverse primers are indicated as white letters on a black background. The nucleotides corresponding to the probe for the target gene from L. interrogans (LI) are highlighted in yellow, while those for the target gene from L. biflexa (LB) are highlighted in green.